The PDB repository and file format#
The Protein Data Bank (https://www.rcsb.org) is the main database holding structural data but there are different entry points into PDB, including the PDB europe (https://www.ebi.ac.uk/pdbe).
The PDB website allows you to search for protein structure information in different ways. You can do simple text queries for names or IDs of proteins or more advanced searches where you can restrict the search to specific species, types of experiments, the quality of the structure, the year it was produced, etc. It is also possible to search by sequence which will return you the structures of sequences that are very similar to the one provided.
As an example, we can find protein structures of sequences that are similar to the protein sequence for the human cAMP-dependent protein kinase (PRKACA). As discussed previously, you can find protein sequence data at the Uniprot database. Searching for human PRKACA leads you to a page where you can retrieve the protein sequence in fasta format.
>sp|P17612|KAPCA_HUMAN cAMP-dependent protein kinase catalytic subunit alpha OS=Homo sapiens OX=9606 GN=PRKACA PE=1 SV=2
MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVML
VKHKETGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMV
MEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY
IQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF
ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFAT
TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF
The protein structure information for PRKACA is also already available in the same Uniprot webpage but we can pretend maybe that this was not the case because your particular protein sequence did not have a structure but there was a structure of homolog that would still be useful for your study.
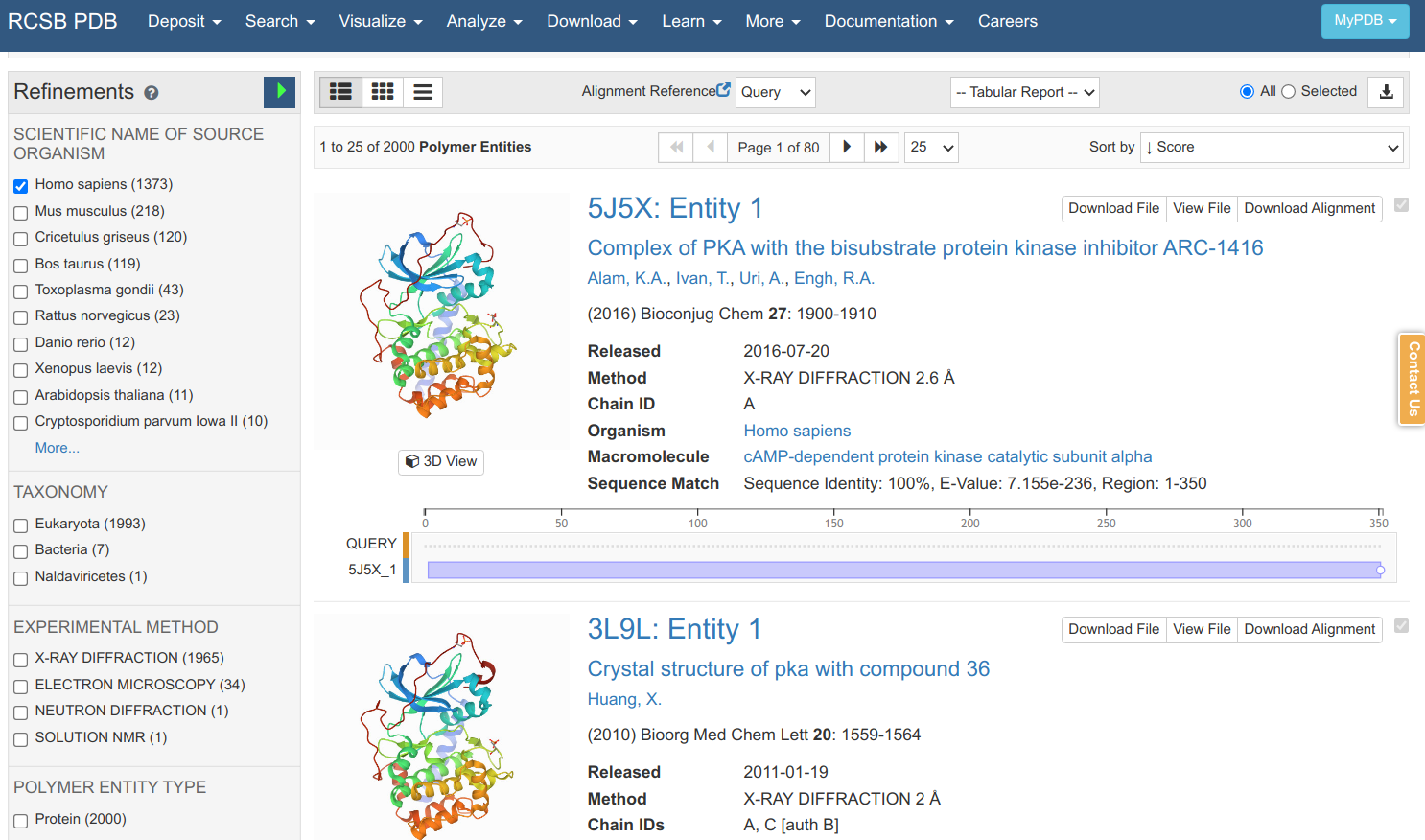
Searching in the PDB with that sequence retrieves many matches that can be further filtered according to the species, experimental method, structural resolution, etc. As it is a sequence search, it also shows you the sequence identity, E-value and how much the protein sequence is covered by the structural model. Clicking on the 5J5X entry leads to an entry specific page.
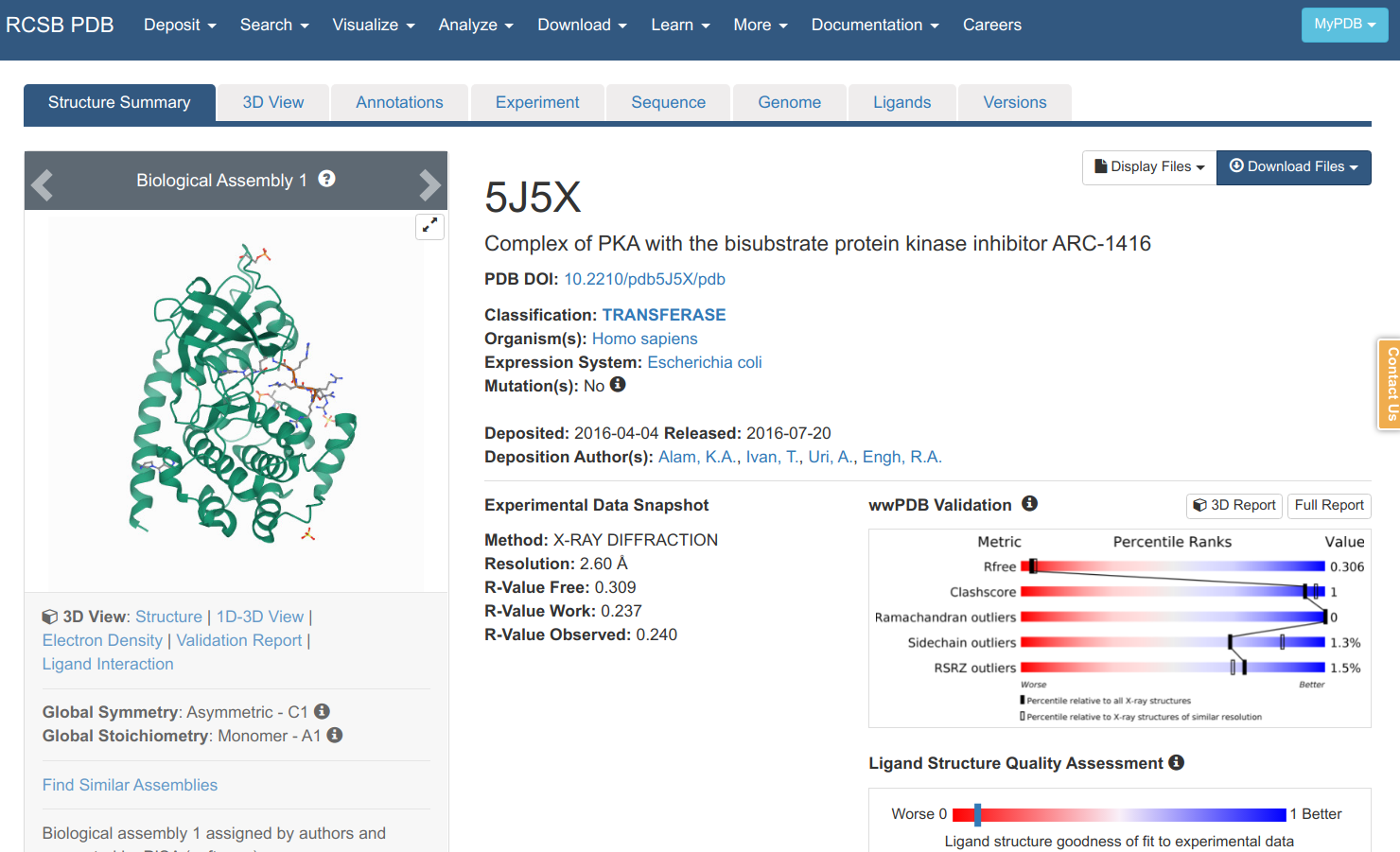
Here you can find information on the experiment, the publication describing the model (if any), and other information on the protein, small molecules etc. You can view a model of the structure by clicking “3D viewer” and retrieve a file that contains the structural information via “Download Files”. There are 2 main file formats that are used to communicate structural data in a standardized way: 1) the PDB format which is older and more human readable and 2) PDBx/mmCIF. If we download the PDB file format and open it in a text editor you can find the most important information for the lines starting with ATOM.
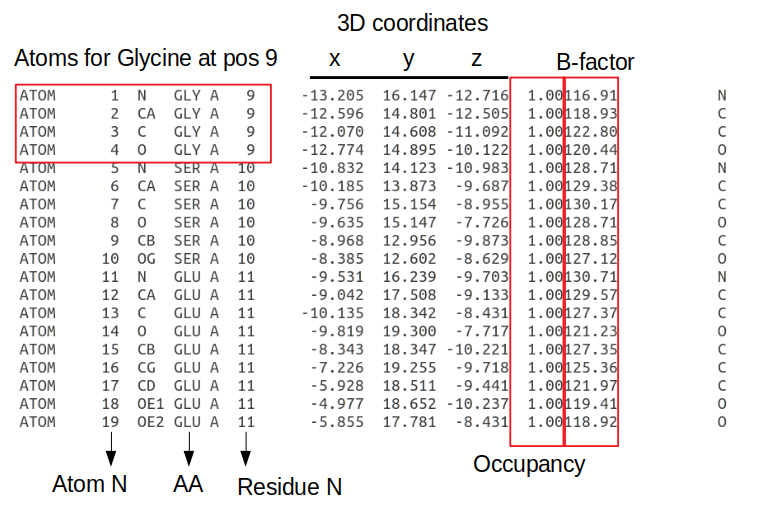
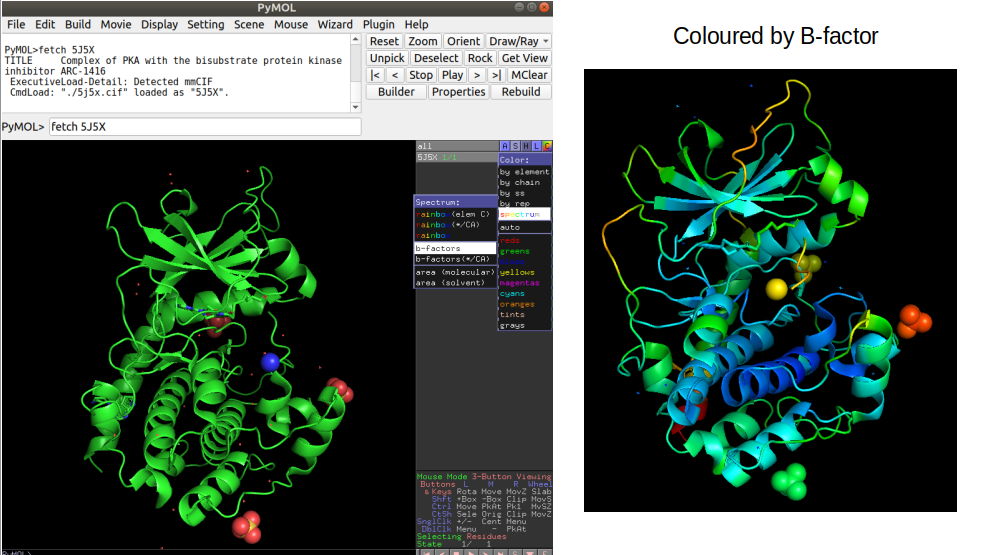
The information is written in a fixed length format where each information type is always at a specific number of characters away from the start of the line. Each line starting with ATOM has information about an atom including the atom number; the atom type (e.g. CA - the alpha carbon of the amino-acid); the amino-acid in a 3 letter code; the chain, the amino-acid residue in the protein sequence; the x/y/z 3D coordinates in angstroms; the occupancy (the fraction of molecules that have each of the conformations); the B-factor or temperature factor (the displacement of the atomic positions from an average value); and again the element name in a one letter code (e.g. C - carbon). The B-factor reflects to some extent how dynamic the residue is with higher numbers representing positions that are less constrained.
In order to visualize a structure Pymol is a useful tool that can be used to perform also some computational operations on selected examples. In pymol, it is possible to load a structure directly by typing “fetch” followed by the PDB id code. For example “fetch 5J5X” will load the PRKACA structure described above. It is possible to color the structure by the B-factor information which illustrates in this case how the lower B-factor scores (blue colors) tend to be within the core of the protein that should be less dynamic. It is important to become familiar with looking at protein structures in order to be critical about the outcome of computational analyses.