Sequence databases#
In your future work, you might want to reference the genome or some of the genes of the organism you are working with, or those of species it is related to. If you generate sequence data, you might want to identify or annotate those sequences using bioinformatic methods that rely on an evidence base of existing public sequence data. It is therefore important that you are aware of the available databases that you might browse or search for such information.
There are three primary sequence databases that essentially contain the same data, exchanged daily between them.
There are additionally a vast array of secondary databases, often specialising in particular types of sequence or individual organisms. We will discuss some of them in future parts of the course.
NCBI#
The National Center for Biotechnology Information (NCBI) hosts a series of databases and tools that are considered essential for modern biology.
The NCBI homepage (below) is a bit overwhelming. At the top you have the search bar (red frame). You can either search (yellow frame) in all databases or you can select a specific database out of the 39 available databases (blue frame).
In the bottom half of the page you have some popular resources on the right side (purple frame) and on the left hand side (green frame) you find a variety of sub areas. In the middle (pink frame) other common features are linked.
In the following section we will describe certain parts of the NCBI to help you find what you are looking for.
NCBI GenBank#
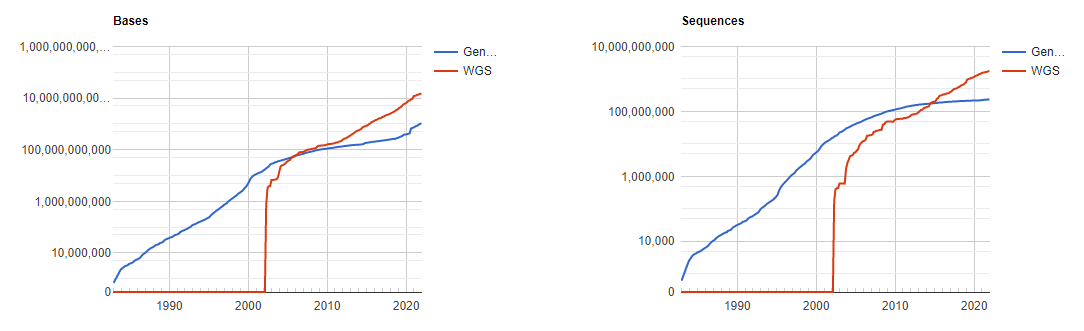
GenBank is an annotated collection of all publically available DNA sequences. This includes genomes, individual genes or feature sequences, transcripts and more. Sequences shorter than 200bp, that aren’t based on a real molecule (for instance a consensus sequence) or that are not known in nucleotide space (for instance a directly sequenced protein), primers, and mixed DNA/mRNA sequences are not accepted. Additional to GenBank is the WGS (whole genome shotgun) database, which contains sequencing projects that are currently the most common form of high-throughput sequencing, but are not yet assembled, finished or annotatable. The graphs below show how the databases have grown over time in number of entries and total base pairs.
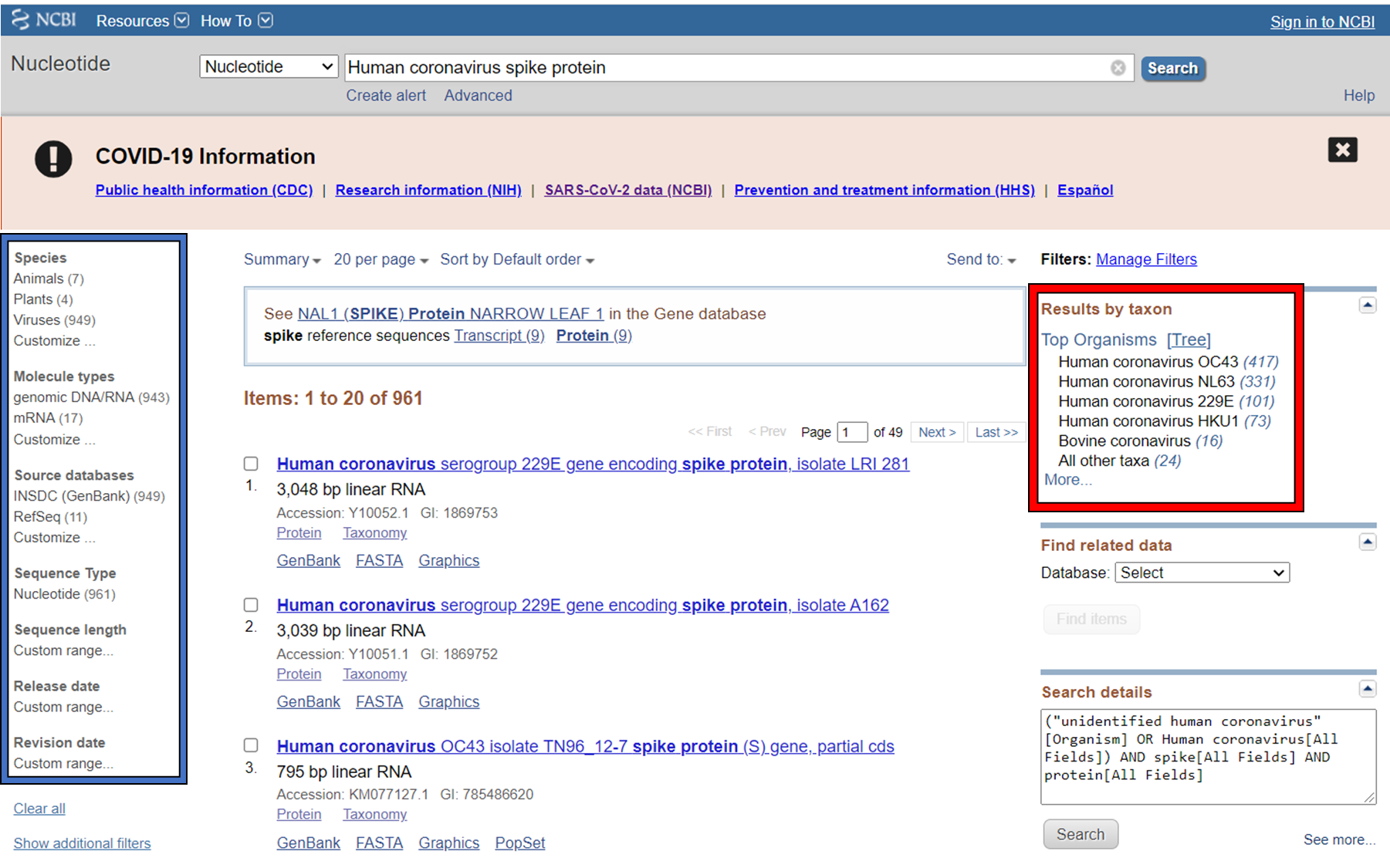
GenBank is searchable by selecting the ‘Nucleotide’ database on the NCBI homepage. It can also be searched by alignment, which will be covered in the next lecture. When you search, you are shown the results as seen below. These can be further filtered by convenient links on the left side of the page (blue frame), or by organism on the right side of the page (red frame).
NCBI RefSeq#
The Reference Sequence database aims to be a comprehensive, well-annotated, non-redundant set of sequences - effectively a curated subset of GenBank to represent the best quality information available for use in biological research. For instance, RefSeq contains 66,541 bacterial entries as of release 2007. If you are looking for a high quality and trustworthy sequence for your work, RefSeq is a good place to start.
RefSeq is not searchable from the NCBI frontpage. Instead, you can search GenBank by selecting the ‘Nucleotide’ database and then use the appropriate filter.
NCBI Genome#
The genome database is another subset of GenBank that includes genomes, chromosomes and assemblies. It aims to assign taxonomy to each entry and give an assessment of completeness. It can be searched directly from the NCBI frontpage by selecting ‘Genome’.
NCBI Taxonomy#
The taxonomy database is a curated classification of the organisms in GenBank, by which we mean their locations on the tree of life. There are alternative taxonomies available, such as the GTDB, as phylogenetic methods differ. Taxonomy is continually under revision, and often submissions are unintentionally misassigned, so be wary when working with less well researched organisms or environments.
Taxonomy can be searched directly from the NCBI frontpage by selecting ‘Taxonomy’.