Structure based prediction of the impact of protein mutations#
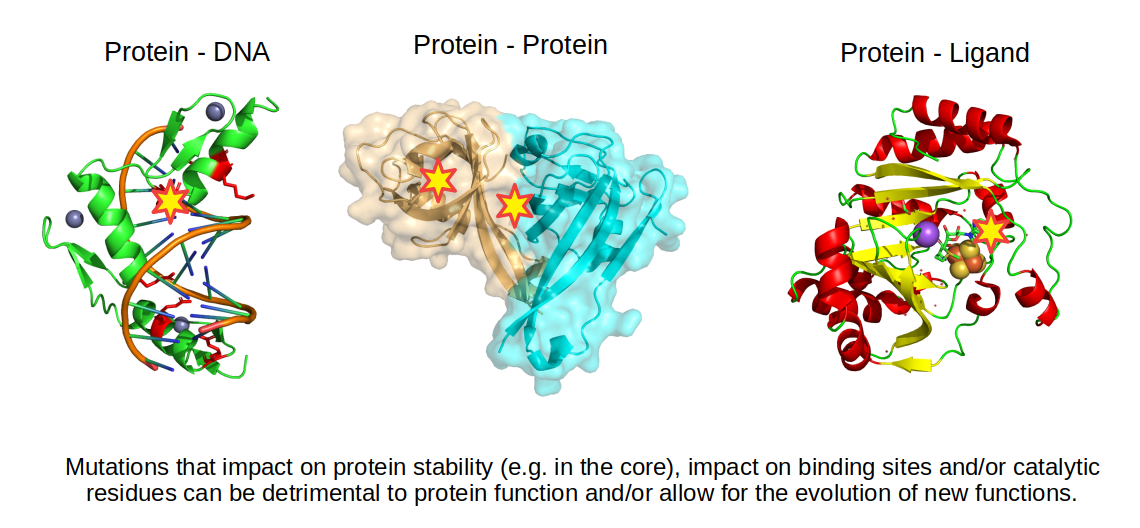
Protein structures are very useful in predicting the impact of a mutation since we can identify changes in amino-acids that could stabilize or destabilize different types of bonds. For example, if a positive residue is making an ionic bond with a negative residue, mutating this negative residue to a positive one will be detrimental to the stability of the structure. Changing residues in the core of the protein to more hydrophilic residues or mutating a small amino-acid to a large amino-acid in the core of the protein may also reduce the stability of the protein. Not all mutations are detrimental, some may change the function of the protein by changing the way the protein interacts with other molecules.
Folding of a protein sequence into a folded state should be thermodynamically stable and the change - or delta - in Gibbs free energy (𝚫G) should be negative when the folded state is more stable than the denatured state. Computationally it is possible to predict this 𝚫G by accounting for all bonds forming within the structure. In addition, the impact of a mutation can then also be predicted and it is often described as the change in 𝚫G or 𝚫𝚫G. When a mutation is destabilizing, 𝚫𝚫G is positive and when it is stabilizing, the 𝚫𝚫G is negative. Most single point mutations in proteins tend not to have an impact on protein stability with 𝚫𝚫G close to 0. Deleterious mutations are strongly enriched when the 𝚫𝚫G >= 2 kcal/mol.
Structural information can also be used to predict the impact of mutations on binding to other molecules. The simplest approach is to find interface residues and overlap with the mutated residues. If the mutation changes dramatically the property of the amino-acid then it may also impact the binding. More sophisticated approaches attempt to quantify the change in binding affinity based on the properties of the bonds formed between the molecules.