Basic file operations#
Besides changing your location on the hierarchical file system, you want to be able to modify the file system itself by creating new directories (folders) or removing empty directories that are not needed anymore. Also, some of the most commonly used commands on any file system are copying, moving and renaming files and removing files that are not needed anymore.
Click on the image below to see what some commands will do within your file system.
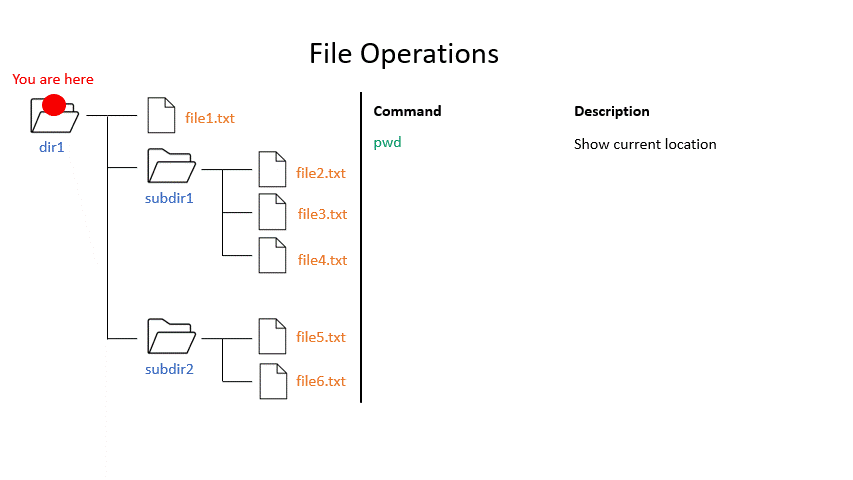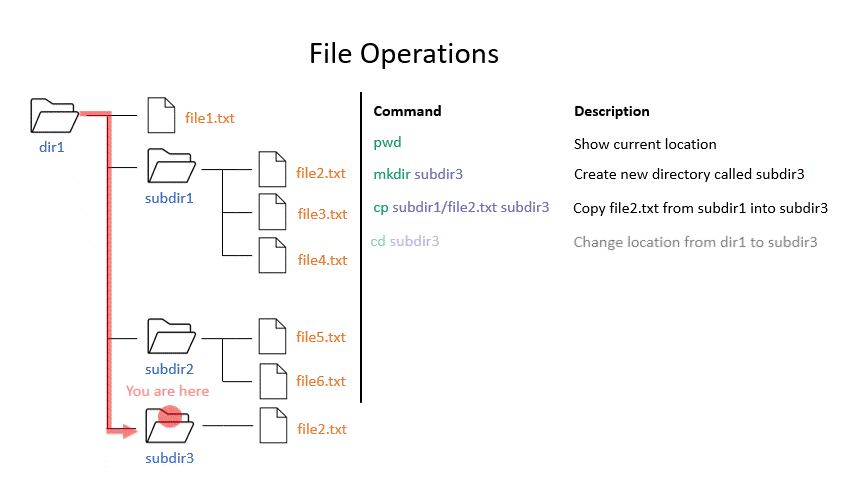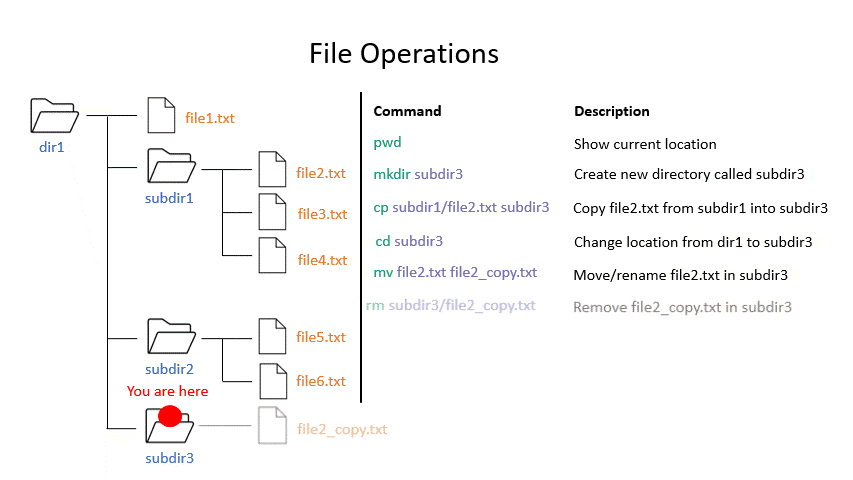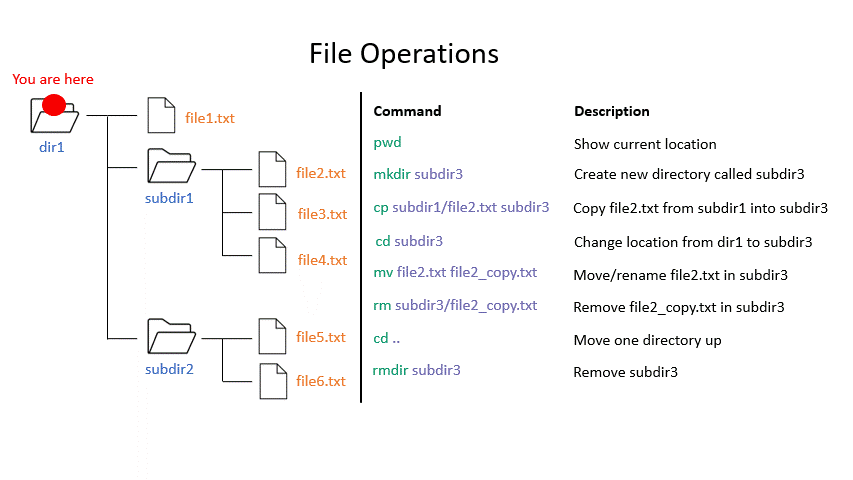
mkdir creates a new directory with the given name.
# Make directory
mkdir <path to directory>
mkdir genomes
rmdir removes an empty directory.
# Remove an empty directory
rmdir <path to directory>
rmdir genomes
cp copies a file from one location to another. The example will copy a file containing the genome sequence of E. coli K12 MG1655 to your home directory.
# Copy
cp <source> <destination>
cp /nfs/teaching/551-0132-00L/1_Unix/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna ~/
mv moves a file from one location to another. The example actually renames the file, because the destination is not a directory. Thus you can move and rename a file with the same command.
# Move or rename
mv <source> <destination>
mv ~/GCF_000005845.2_ASM584v2_genomic.fna ~/E.coli_K12_MG1655.fna
rm removes a file, so use it with care.
# Remove
rm <path_to_file>
rm ~/E.coli_K12_MG1655.fna
Exercise 1.5#
Exercise 1.5
Create three new directories called “genomes”, “test” and “in_class” in your home folder
# First go to your home folder
cd
# Use the mkdir function to create a directory
mkdir genomes
mkdir test
mkdir in_class
Delete the test directory
# Use the rmdir function to remove a directory
rmdir test
Copy the file
/nfs/teaching/551-0132-00L/1_Unix/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna
into your new directory “genomes”
# Use the cp function to copy. cp <source> <destination>
cp /nfs/teaching/551-0132-00L/1_Unix/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna ~/genomes
Rename the file to “E.coli_K12_MG1655.fna”
# Use the move function to rename a file mv <source> <destination>
# Enter the genomes directory
cd genomes
# Rename file
mv GCF_000005845.2_ASM584v2_genomic.fna E.coli_K12_MG1655.fna
Make a copy of the
~/genomes/E.coli_K12_MG1655.fnafile and rename it ‘E.coli_K12_MG1655_Copy.fna’, then delete the copied file
# Use the cp function to copy. cp <source> <destination>
cp ~/genomes/E.coli_K12_MG1655.fna ~/genomes/E.coli_K12_MG1655_Copy.fna
# Here you see that you can use the function cp to copy and rename a file in one step, if you give it a new file name instead of just a desination directory
# Use the functions cd and ls to check whether the renamed file copy is in your ~/genomes directory
cd ~/genomes
ls
# Only after checking, use the function rm to remove the copied and renamed file (pay attention not to remove the original file but the renamed copy)
rm E.coli_K12_MG1655_Copy.fna
Use cp to copy all files from the directory
/nfs/teaching/551-0132-00L/1_Unix/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2
into a new directory in your home directory
# Make a directory for the new files
cd ~
mkdir ecoli
# Copy all the files
cp /nfs/teaching/551-0132-00L/1_Unix/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/* ~/ecoli/
# The character '*' is a wildcard which stands for any number of arbitrary characters following the path of the chosen directory, meaning that all files within that directory are chosen as arguments for the command. If you want to know more, wildcards are described in more details in the additional content.