Phylogenetic trees#
The objectives for reconstructing phylogenetic trees can be manifold. Generally speaking, a phylogenetic tree is a hypothesis of how biological species or other entities (e.g., genes) are related through evolution. It is a branching diagram showing the inferred evolutionary relationships among these entities based on similarities in their genetic and/or physical characteristics.
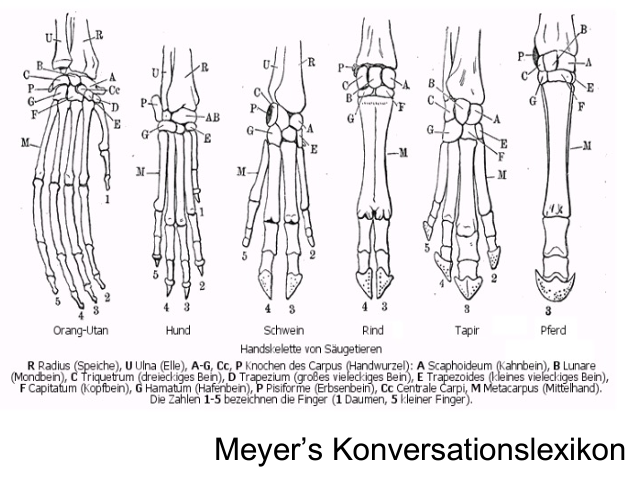
For the interpretation of phylogenetic trees, it is important to understand the concept of homology as similarity due to shared ancestry. For example, the forelimbs of vertebrates are homologous structures. Although in different animals, they may vary in form and function (e.g., arms, forelegs, wings, front flippers), they have evolved from the same structure in the last common ancestor of tetrapods. However, the function of wings in insects, bats and birds is analogous, as it has evolved independently in widely divergent groups of animals.
Below you can see a comparison of forelimbs of different mammals, which are closely related but still contain significant structural differences.
Sequence homology#
By extension of the concept of homology to DNA and protein sequences, two sequences are homologous if they share ancestry. High similarity between two sequences provide strong evidence for their shared ancestry, but is my no means conclusive. Importantly, based on the definition of homology specified above, the similarity between sequences is merely an empirical observation. Whether or not these sequences are homologous requires interpretation, e.g. by reconstructing phylogenetic trees. As with wings, sequence similarity may occur as a result of convergent evolution, or with short sequences, by chance.
Furthermore, homologous sequences can be orthologous or paralogous with respect to each other: “Where the homology is the result of gene duplication so that both copies have descended side by side during the history of an organism, (for example, alpha and beta hemoglobin) the genes should be called paralogous (para = in parallel). Where the homology is the result of speciation so that the history of the gene reflects the history of the species (for example alpha hemoglobin in man and mouse) the genes should be called orthologous (ortho = exact).” – W. Fitch. Homologous sequences that have been transfered between species are xenologs.
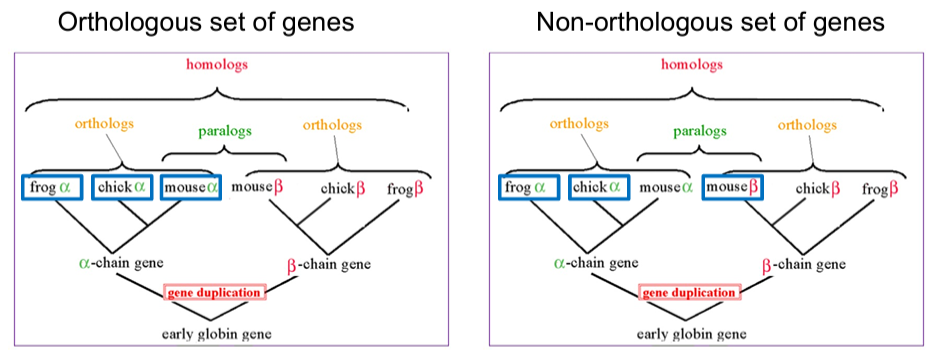
One of the most important implications for phylogenetics is that only sets of orthologous sequences are expected to reflect the underlying evolution of species, whereas a set of homologous genes (including orthologs, paralogs and xenologs) can be informative about the evolutionary relationship between species (gene duplication within/among species and horizontal gene transfer). Orthologous genes, as compared to paralogs, are also more likely to share the same function.