Sequencing-based transcriptomics#
Since being introduced in the min 2000s, RNA-Sequencing has become an almost ubiquitous method for the structural and quantitative assessment of the transcriptome of an organism. Initially prohibitively costly when compared to array-based approaches, sequencing has become an affordable alternative addressing some of the major limitations of microarrays.
There is many online resources available that summarize the technology and related matters at great detail (e.g., the RNA-seqlopedia). We will give a broad overview here and refer to these additional materials for any further details.
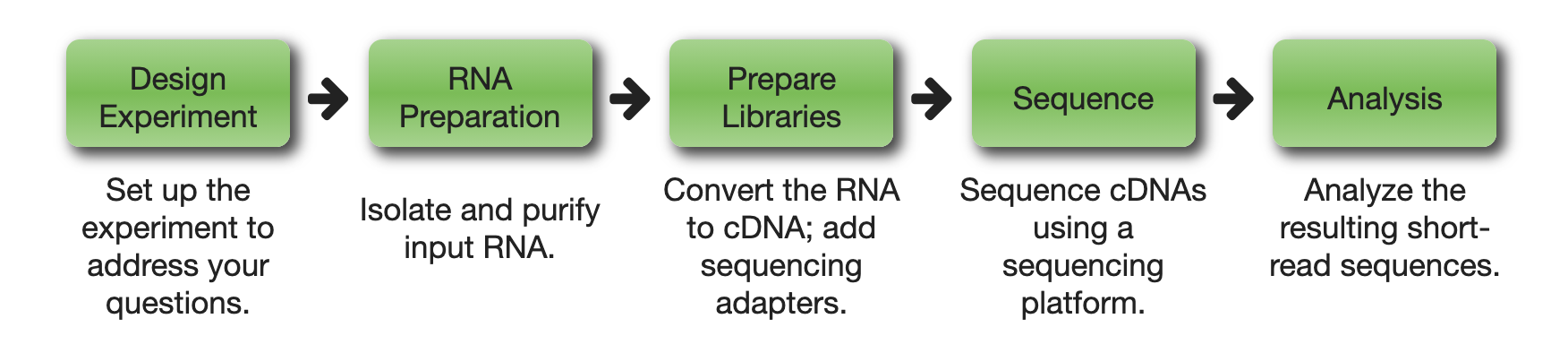
Flowchart outlining the main steps of an RNA-Sequencing experiment. (Source: https://rnaseq.uoregon.edu/#exp-design)#
After experimental design, the RNA-Seq workflow consists of the following steps
1. RNA Isolation and Purification - The biological sample (cells, tissues, or other sources of RNA) re lysed to release the RNA, which is then isolated. Depending on the research goal, specific RNA-subtypes, such as mRNA or miRNA, can be isolated. To purify the RNA, contaminants like DNA and proteins are removed. Common methods include phenol-chloroform extraction or using commercial RNA extraction kits.
2. RNA Quality Assessment - To guarantee integrity of further processing steps, it is essential to assess the quality and quantity of your isolated RNA. Common methods for quality control include using a spectrophotometer (e.g., Nanodrop) and an Agilent Bioanalyzer to assess RNA integrity.
3. RNA Fragmentation - As part of some RNA-Seq protocols, the RNA is fragmented to a desired size range (e.g., 200-300 base pairs) using heat or enzymatic fragmentation. The exact method may vary depending on the specific RNA-Seq library preparation kit being used.
4. cDNA Synthesis - In the first step of reverse transcription the fragmented RNA is converted into complementary DNA (cDNA) using reverse transcriptase and random hexamer. oligo(dT) or other specific primers. oligo(dT) primers would preferentially select polyadenylated mRNA from the sample. After reverse transcription, second strand synthesis generates a second strand of cDNA, effectively creating double-stranded cDNA.
5. End Repair and A-Tailing - The cDNA fragments from the previous step are subjected to end repair, where overhangs are removed, and polyA-tails are added. This step prepares the ends for adapter ligation.
6. Adapter Ligation - Adapters are short sequences containing information necessary for sequencing (such as barcodes or indices for multiplexing). They are ligated to the A-tailed cDNA fragments. This step allows the sequencer to identify and demultiplex samples.
7. Size Selection and Purification - The cDNA fragments with ligated adapters are size-selected to enrich for fragments of the desired size range. This is often done by gel electrophoresis or through mechanical beating with magnetic bead.
8. Amplification (Optional) - In most sequencing protocols, cDNA fragments are amplified via PCR to increase the amount of material for sequencing. If sufficient amount of input material is available no amplification is necessary.
9. Library Quality Control - The product of the above steps is called a sequencing library. Assessing the quality of the prepared library includes checking the size distribution of cDNA fragments and quantifying the library using methods like qPCR or fluorimetry.
10. Sequencing - The prepared library is loaded onto a high-throughput sequencing platform such as Illumina, Ion Torrent, or Oxford Nanopore sequencers. The sequencer reads the DNA sequences of the cDNA fragments, generating raw data in the form of short (Illumina) or long (ONT) reads.
11. Data Analysis - After sequencing, the raw data is processed through a series of bioinformatics analysis steps, which can include read mapping, transcript assembly, differential gene expression analysis, and various downstream analyses to derive meaningful biological insights.
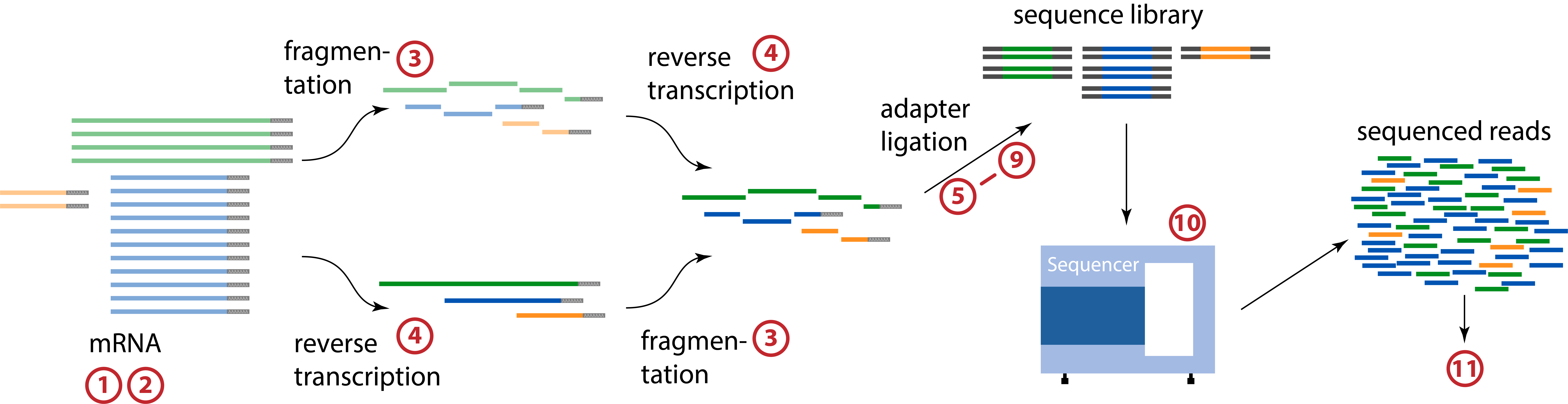
Schematic approximation of the above RNA-Seq process.#