R-Studio#
Various parts of the Bioinformatics Concepts Course will be based in R-Studio. Instead of relying on a local installation on your computer, we have made R-Studio easily accessible to you on our server. This R-Studio has already been set up with all the necessary packages that you will need during the course. Click on the link below to load R-Studio in your browser:
R-Studio: http://cousteau-rstudio.ethz.ch/
Note
You need to use the web-based R-Studio on our server - your local version may not have the correct packages or set up for the course.
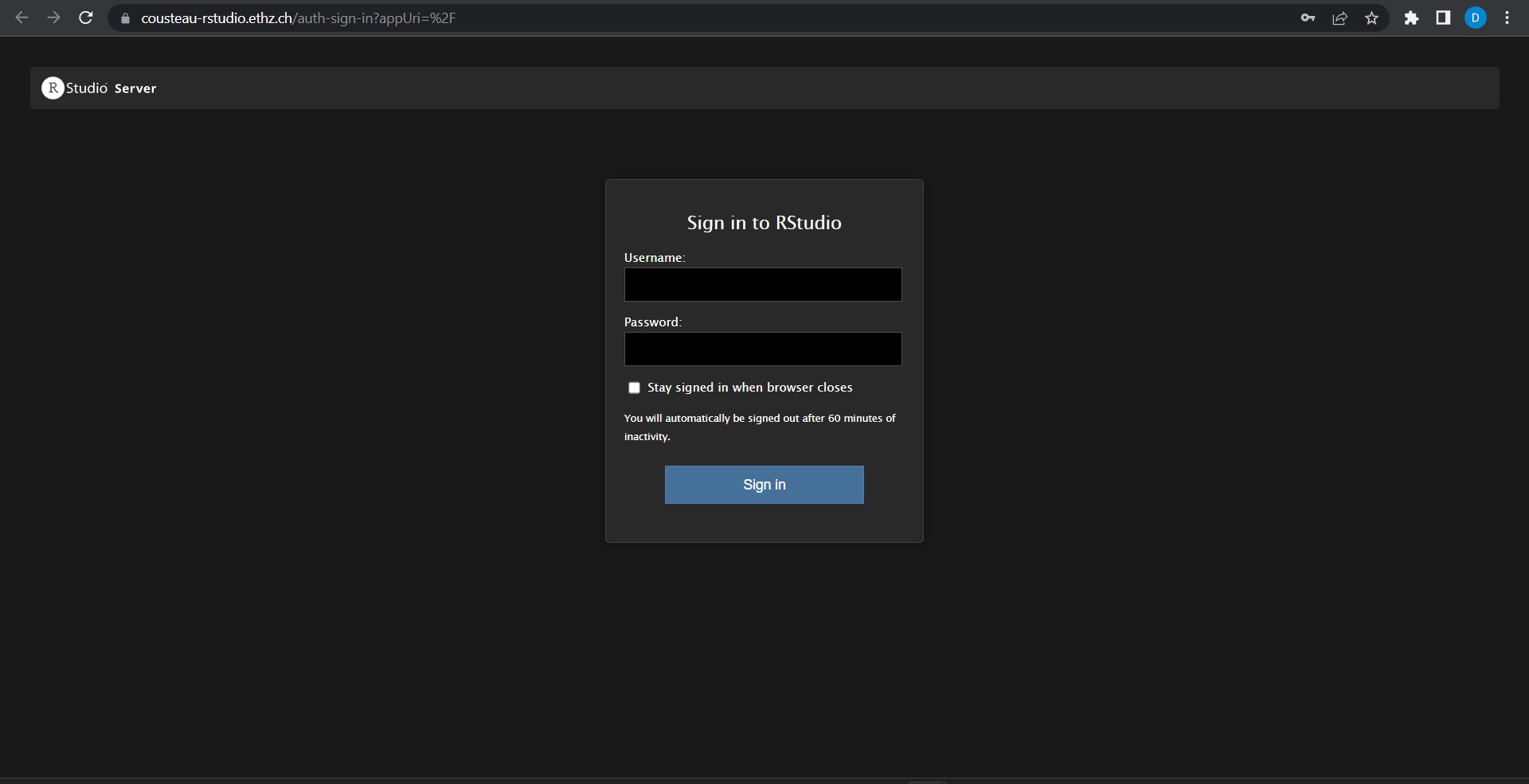
After clicking on the link, a login screen should appear (picture below). You can use your normal ethz-login, which will then provide you access to the R-Studio.
Note
Due to technical reasons, it is possible that you have to login twice when you first try to connect to R-Studio on our server.
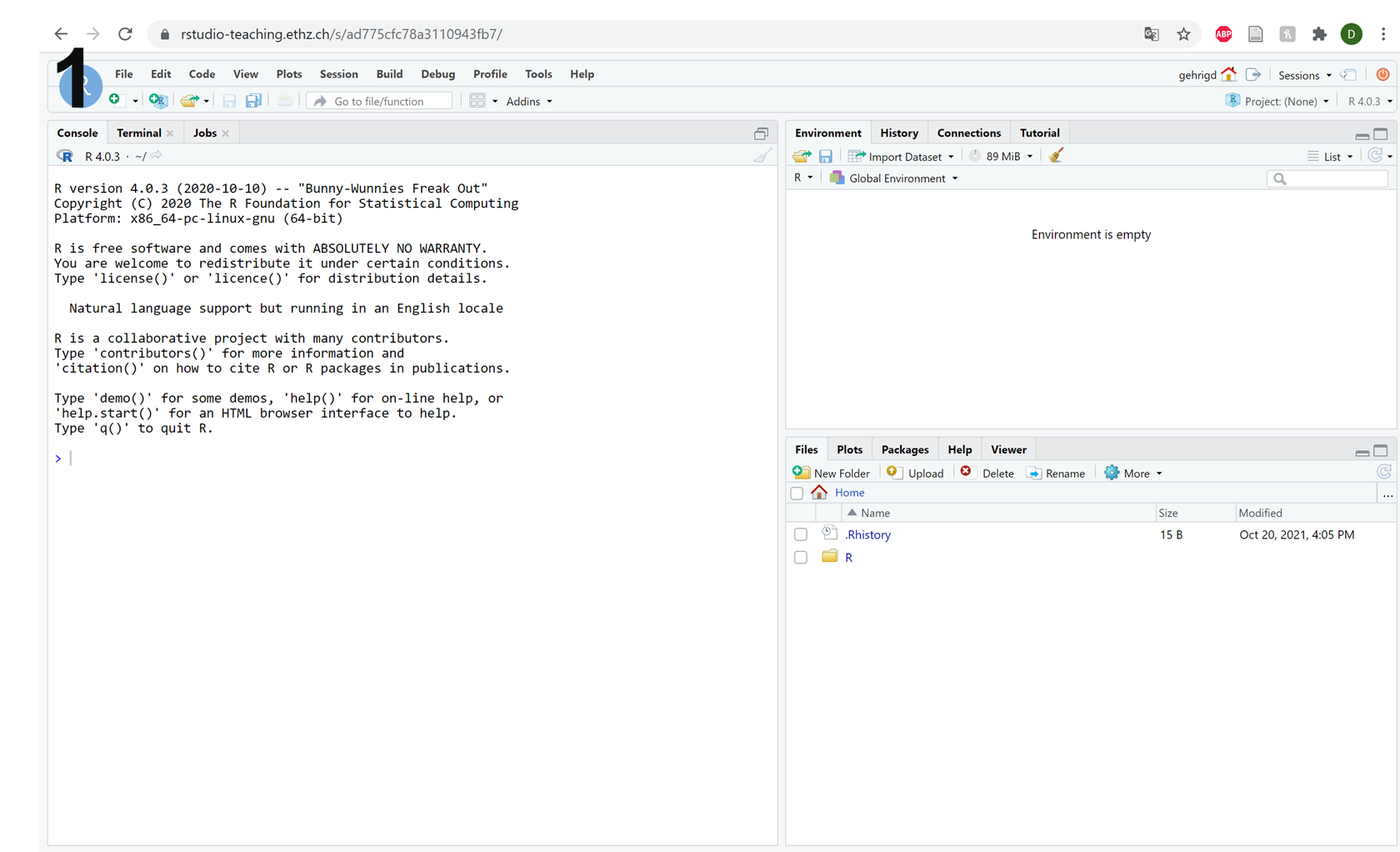
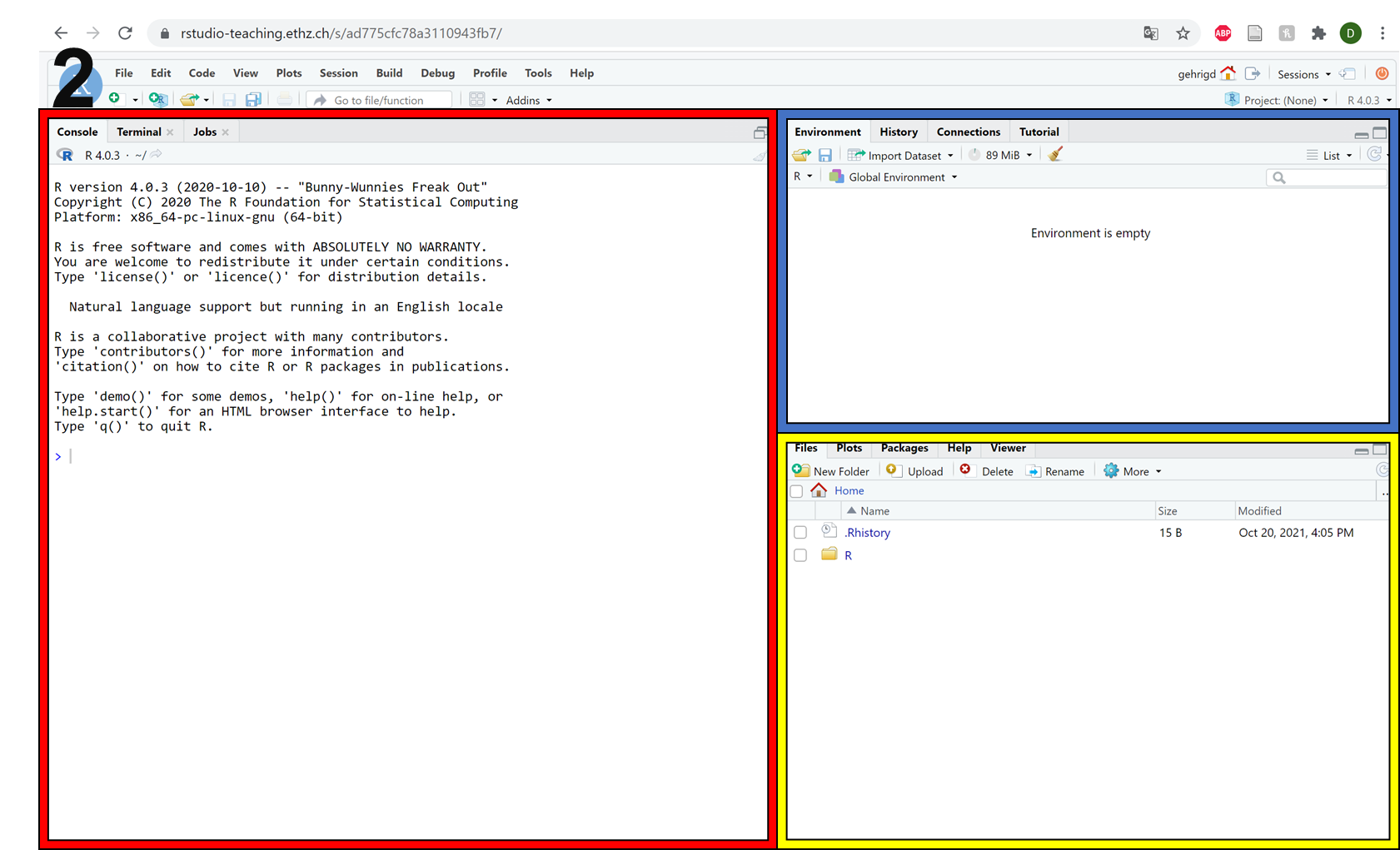
After successfully logging in, you will be presented with the standard R-Studio interface (picture 1 below). Hopefully, you are familiar with this interface. As a brief reminder, there are three main sections (picture 2 below), which correspond to:
The red frame shows the R console. Here, you can write and run R code and see the results.
The blue frame shows the environment. Here, you will find all of the variables that you have created and any datasets that you have loaded.
The yellow frame shows your home folder on our server by default. However, you can toggle across the different tabs to view the ‘packages’ that you have loaded, or the ‘plots’ that you have created in your current R session.
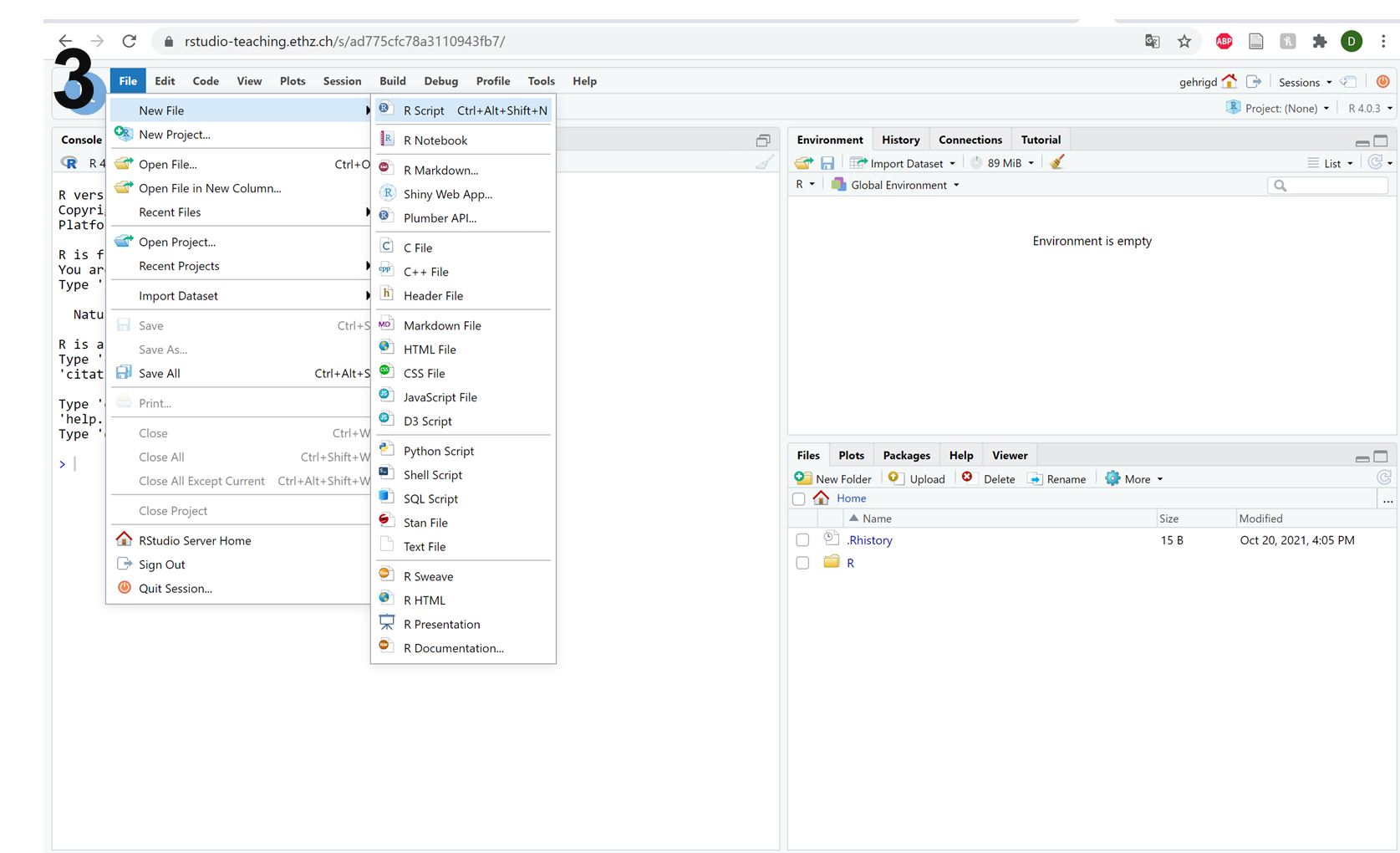
To complete the set-up and begin working on course material, you will need to create a new R-script (Picture 3 below): Click on File -> New File -> R Script (or use the shortcut with key combination Ctrl + Alt + Shift + N) as you can see in picture 3.
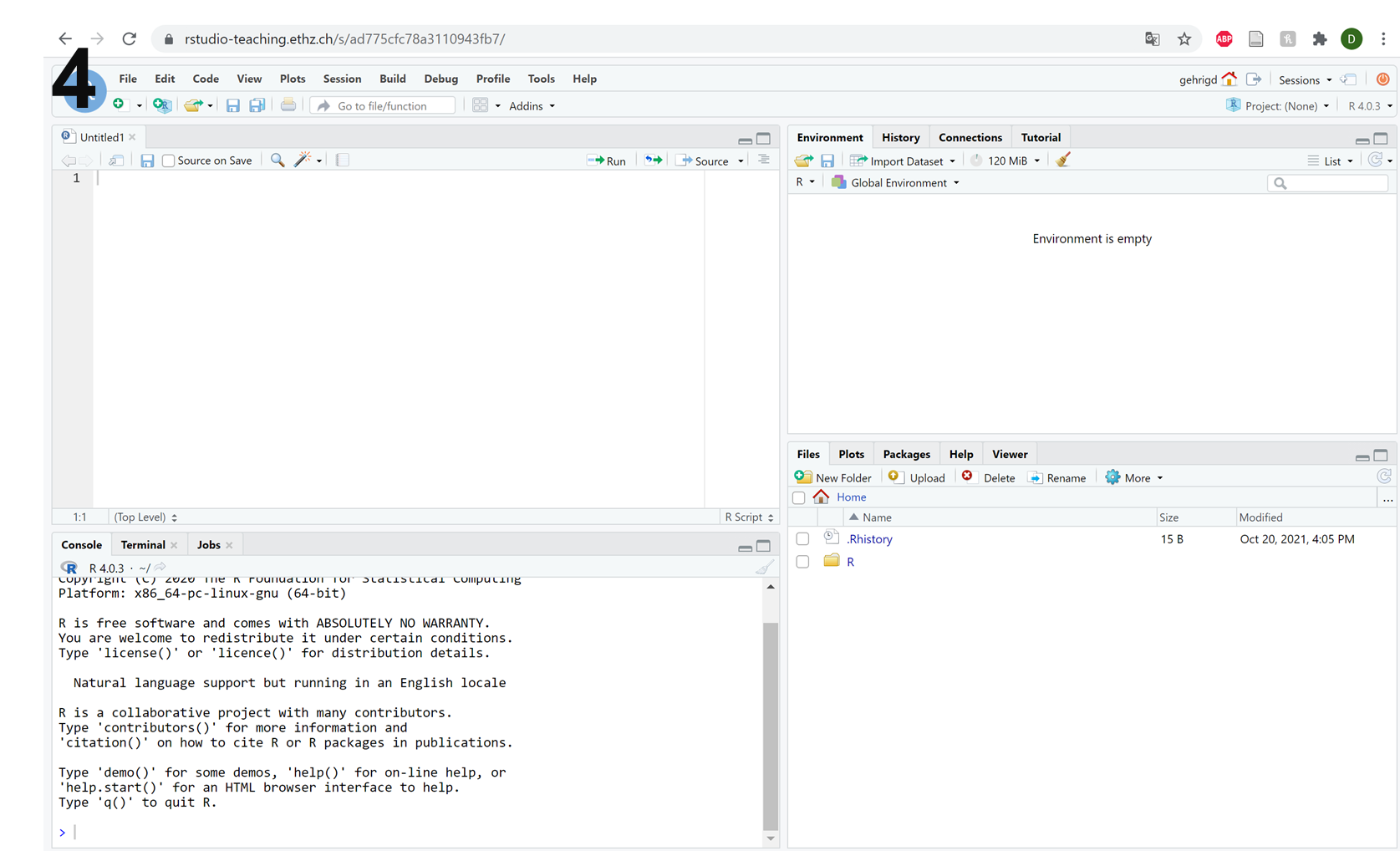
Following this, you should have four panels in your R-Studio interface (picture 4 below), at which point the set-up is complete!