Basic file operations#
cp copies a file from one location to another. The example will copy a file containing the genome sequence of E. coli K12 MG1655 to your home directory.
# Copy
cp <source> <destination>
cp /nfs/teaching/551-0132-00L/1_Unix/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna ~/
mv moves a file from one location to another. The example actually renames the file, because the destination is not a directory. Thus you can move and rename a file with the same command.
# Move or rename
mv <source> <destination>
mv ~/GCF_000005845.2_ASM584v2_genomic.fna ~/E.coli_K12_MG1655.fna
rm removes a file, so use it with care.
# Remove
rm <path_to_file>
rm ~/E.coli_K12_MG1655.fna
mkdir creates a new directory with the given name.
# Make directory
mkdir <path to directory>
mkdir genomes
rmdir removes an empty directory.
# Remove an empty directory
rmdir <path to directory>
rmdir genomes
Click on the image below to see what every command will do within your file system.
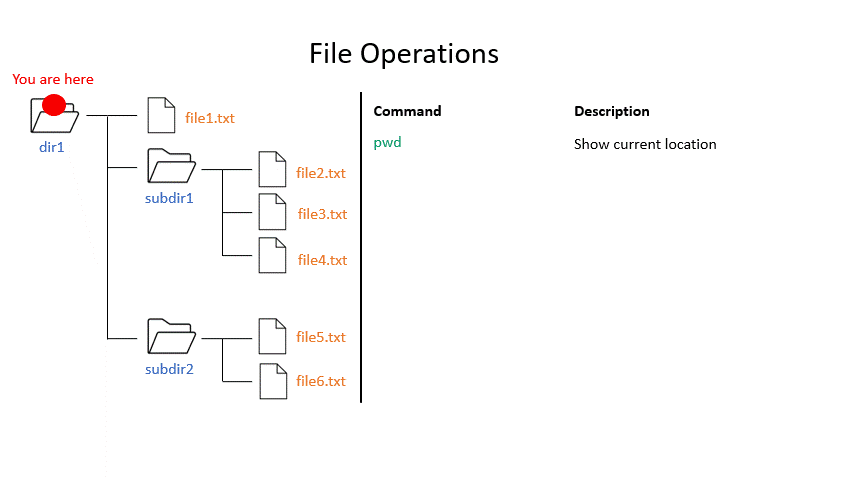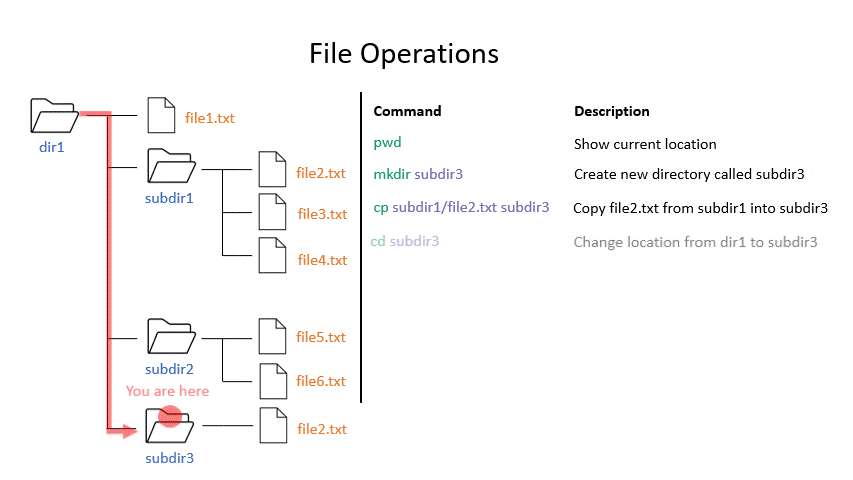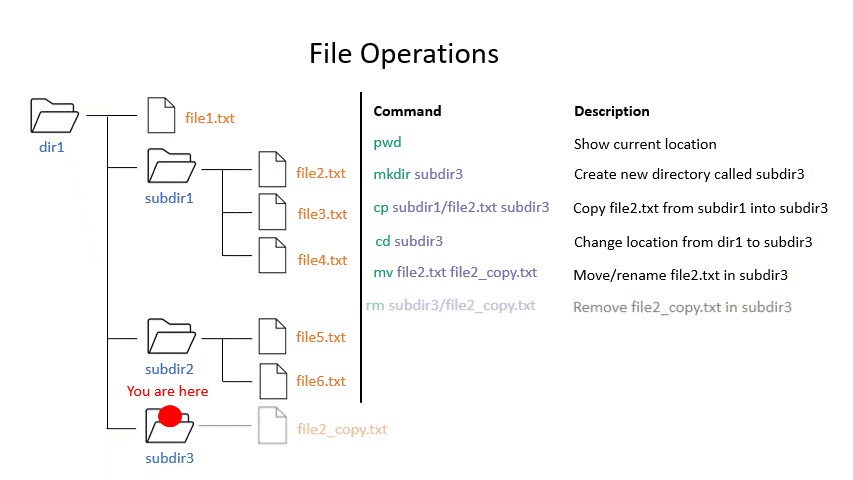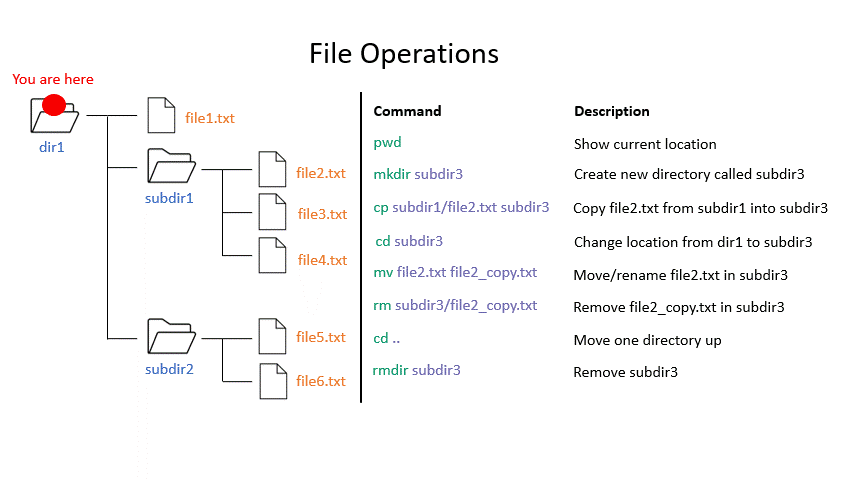
Exercise 0.3
Create three new directories called “genomes”, “test” and “in_class” in your home folder
# First go to your home folder
cd
# Use the mkdir function to create a directory
mkdir genomes
mkdir test
mkdir in_class
Delete the test directory
# Use the rmdir function to remove a directory
rmdir test
Copy the
/nfs/teaching/551-0132-00L/1_Unix/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fnafile into your new directory “genomes”
# Use the cp function to copy. cp <source> <destination>
cp /nfs/teaching/551-0132-00L/1_Unix/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna ~/genomes
Rename the file to “E.coli_K12_MG1655.fna”
# Use the move function to rename a file mv <source> <destination>
# Enter the genomes directory
cd genomes
# Rename file
mv GCF_000005845.2_ASM584v2_genomic.fna E.coli_K12_MG1655.fna
Use the help option of the ls function to find out which option will give file size in a human-readable format. Remember that ls -l will give a table of file information including size.
# ls -l will give you a table of information but the file sizes are long and hard to read
ls -l
# ls --help lists all the options possible
ls --help
# The -h option makes file sizes human-readable, however file sizes are only printed when you use the -l flag so you must use both
ls -l -h
ls -lh
# The size should be 4.5M
Using man and cp, find out how to copy a directory.
# Enter home directory
cd
# Create two directory
mkdir dir1
mkdir dir2
# Try to copy dir1 into dir2
cp dir1 dir2
cp: dir1 is a directory (not copied).
# If you check 'man cp', you see that you have to use -R:
man cp
cp -R dir1 dir2
# Check if the directory has been copied
ls dir2
File name conventions#
In Unix systems there are only really two types of files: text or binary. The file name ending (.txt or .jpg) doesn’t really matter like it does in Windows or Mac OS, however it is used to indicate the file type by convl encounter include:pes you will
.txt - A generic text file.
.csv - A ‘comma separated values’ file, which is usually a table of data with each line a row and each column separated by a comma.
.tsv - A ‘tab separated values’ file, which is the same but separated by tab characters.
.fasta or .fa - A fasta formatted sequence file, in which each sequence has a header line starting with ‘>’.
.fna - A fasta formatted nucleotide sequence file, usually gene sequences.
.faa - A fasta formatted protein sequence file.
.sh - A ‘shell script’, which contains commands to run.
.r - An R script, which contains R commands to run.
.py - A python script, which contains python commands to run.
.gz or .tar.gz - A file that has been compressed using a protocol called ‘gzip’ so that it takes up less space on the disk and transfers over the internet faster.