Sequencing Technologies#
In the following, we will provide a brief historic overview of the different generations of sequencing technologies.
Sanger sequencing#
Proposed in 1977 by Frederick Sanger, this technology is still used today due to its high accuracy and relatively long read length.
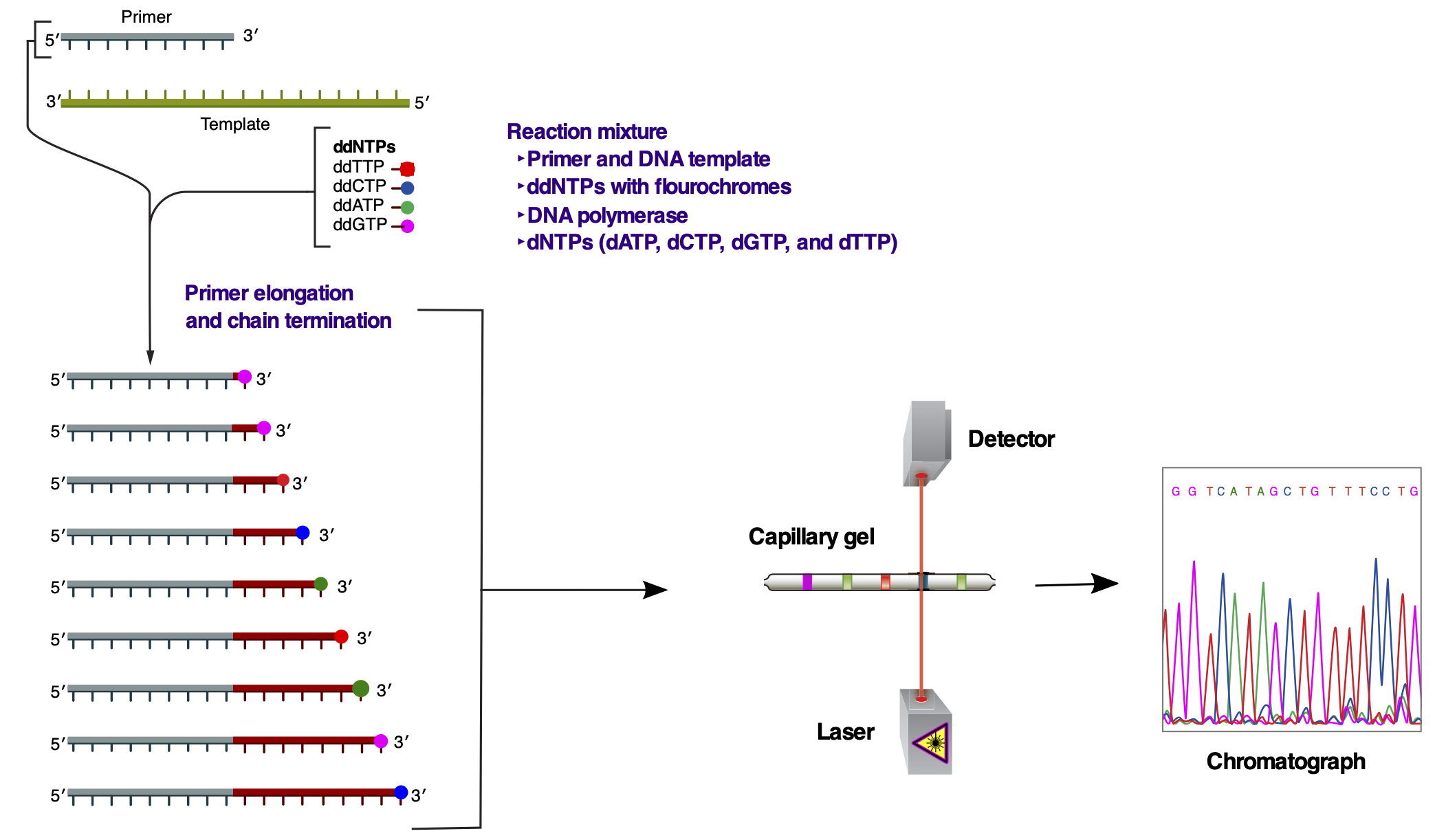
Overview of the Sanger sequencing strategy. (This image has been modifed from https://en.wikipedia.org/wiki/Sanger_sequencing#/media/File:Sanger-sequencing.svg)#
The general idea of the Sanger sequencing technology is to exploit base-complementarity of DNA (or RNA) to synthesise the remaining strand from a given single-stranded molecule (the template). The template is already paired with a primer sequence, forming a short stretch of double-stranded molecule, serving as the origin for sequence extension.
The reaction mix contains both deoxynucleotide triphospates (dNTPs) as well as dideoxynucleotide triphospates (ddNTPs). The latter prevent any further extension of the molecule. Mixing both dNTPs and ddNTPs for sequencing will cause the sequences resulting from elongation to have a spectrum of different lengths. In addition, each ddNTP is attached to a distinct fluorochrome that allows for optic detection. Sorting the synthesised sequences by size and reading out the fluorescence information will then generate the sequence of the originally provided template molecule.
Originally the size sorting was done using a DNA gel and the readout was performed manually. Since the 1990s, the size sorting is done using capillary gels and the scanning of fluorescence signal is automated.
The Sanger technique is highly accurate (with an error rate of less than 0.01%) and produces reads with a length of 500-1000 nucleotides.
- Key characteristics:
very low throughput (single sequences)
very high accuracy (< 0.001% error rate)
moderate read lengths (500-1000 nucleotides)
Illumina sequencing#
Introduced in 1998 by different companies, the “second generation” sequencing technologies follow the shotgun principle. In this strategy the DNA to be sequenced is sheared into smaller fragments which are then sequenced in a highly parallel manner. Also this technology follows the sequencing-by-synthesis (SBS) paradigm, where the complementarity of DNA is exploited to generate sequence based on a given template strand.
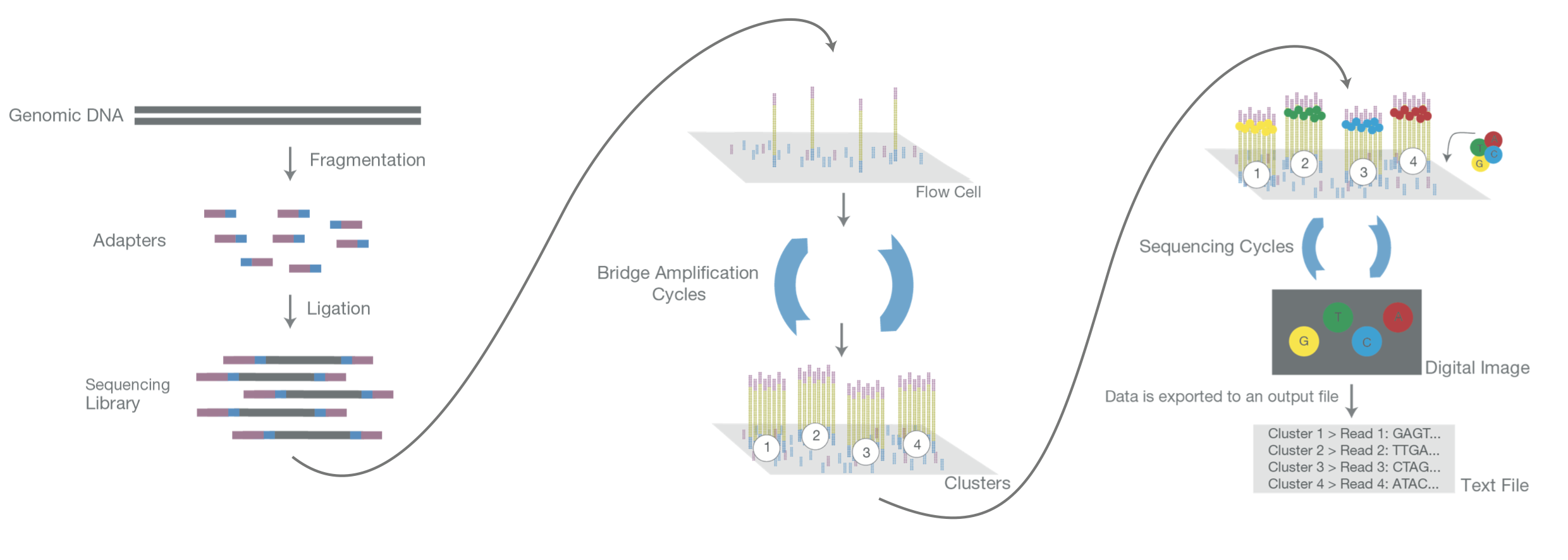
Overview of the Illumina sequencing strategy. (This image has been modifed from https://emea.illumina.com/content/dam/illumina-marketing/documents/products/illumina_sequencing_introduction.pdf)#
After shearing the input DNA into short fragments (usually 400-1000 nucleotides in size), sequencing adapters are fused to the molecules. (If too little input DNA is available, the fragments can be amplified using the polymerase chain reaction (PCR) prior to adapter ligation.) The sequencing adapters allow the fragments to bind to short oligo-nucleotides at the bottom of the sequencing flow cell. Once bound to the flow cell, the molecules undergo a further step of local amplification that increases their number exponentially. This step is also called clustering and the resulting local spots of identical molecules sequence clusters.
Subsequently, many rounds of simultaneous sequence extension are performed where in each round one of the four nucleotides (A, C, G, T) is added. The newly added nucleotides are fused to a fluorophore that allows measuring which sequence-clusters are currently being extended. From the stack of images, where each image depicts the current extension of mullions of different read clusters, the sequence of each template in each spot can be computationally determined.
Key characteristics:
high throughput (~ 65 Gb / hour)
low error rate (< 0.01 %)
short read lengths (50-250 nt; can be paired-end, sequencing both ends of same fragment)
PacBio sequencing#
In 2004 Pacific Biosciences (PacBio; then still under a different company name) introduced the SMRT single molecule sequencing technology - beginning the era of 3rd-generation sequencing technologies.
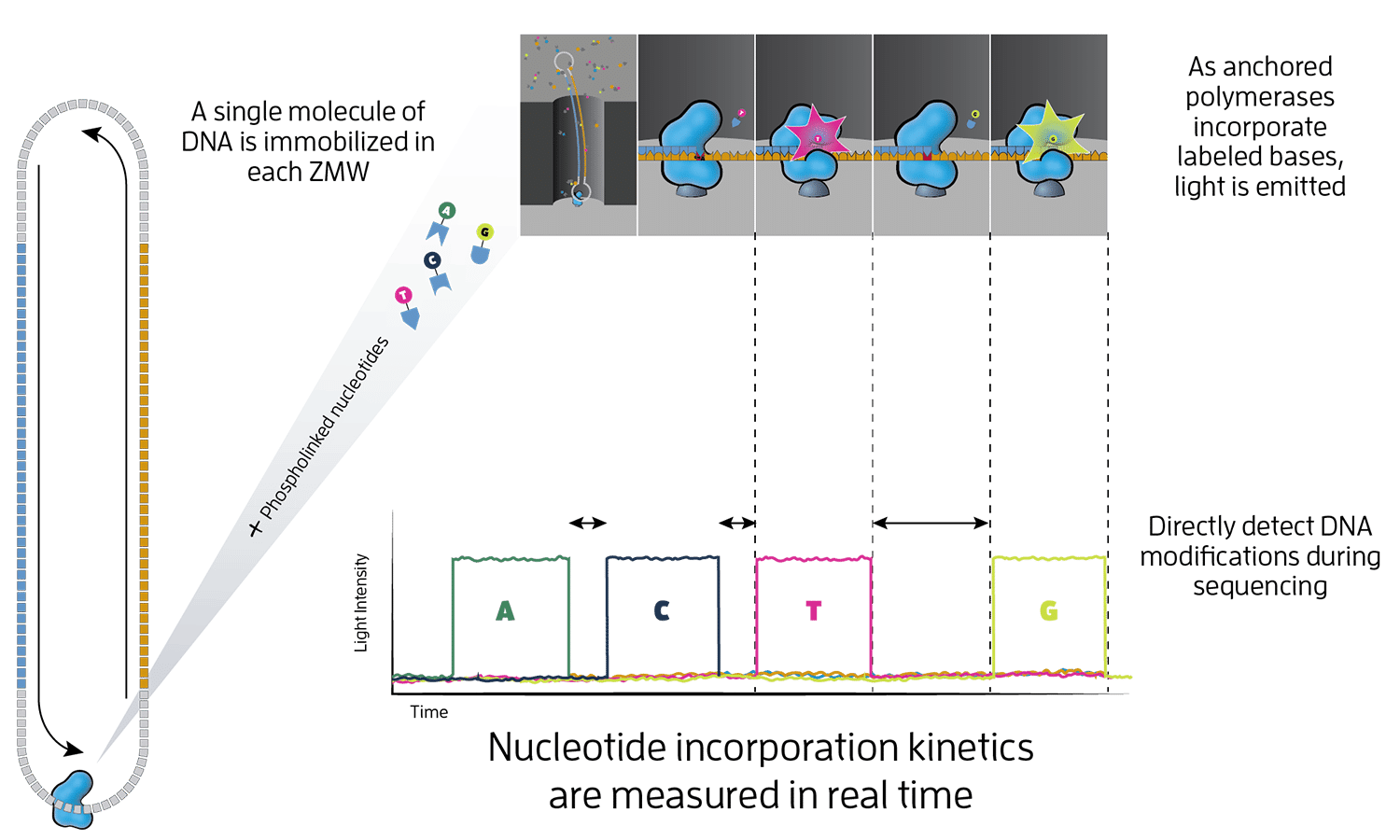
Overview of the PacBio sequencing strategy. (This image has been taken from https://www.pacb.com/wp-content/uploads/Epigenetics.png)#
Here, a single DNA molecule is attached to a polymerase, which is then fixated at the bottom of a nano-well - which is also called a zero-mode waveguide (ZMW). Within the ZMW the polymerase completes the single strand into a double strand, in the process incorporating fluorescence-labelled nucleotides. The labelled nucleotides emit a fluorescence signal during incorporation which is visually recorded and then digitally analysed. From the sequence of recorded signals the base-sequence is computed. As any errors are random, the molecules to be sequenced can be circularised and read multiple times. Hence the same sequence is measured over and over again, lowering the overall error.
As the method is amplification-free, the technology can also detect modifications of the DNA, such as methylations, allowing direct access to epigenomic signals. With measuring many ZMWs in parallel, the technology reaches the required throughput for deep sequencing, with a throughput of up to 360 gb per day for the most recent (Revio) platforms.
Key characteristics:
medium high throughput (~2-15 Gb / hour)
moderate to high error rate (1-10 %)
long read length (median of 15-20 kb)
Nanopore sequencing#
Introduced in 2005, the Oxford Nanopore Technologies (ONT) sequencing technology is the second prominent representative of 3rd-generation sequencing.
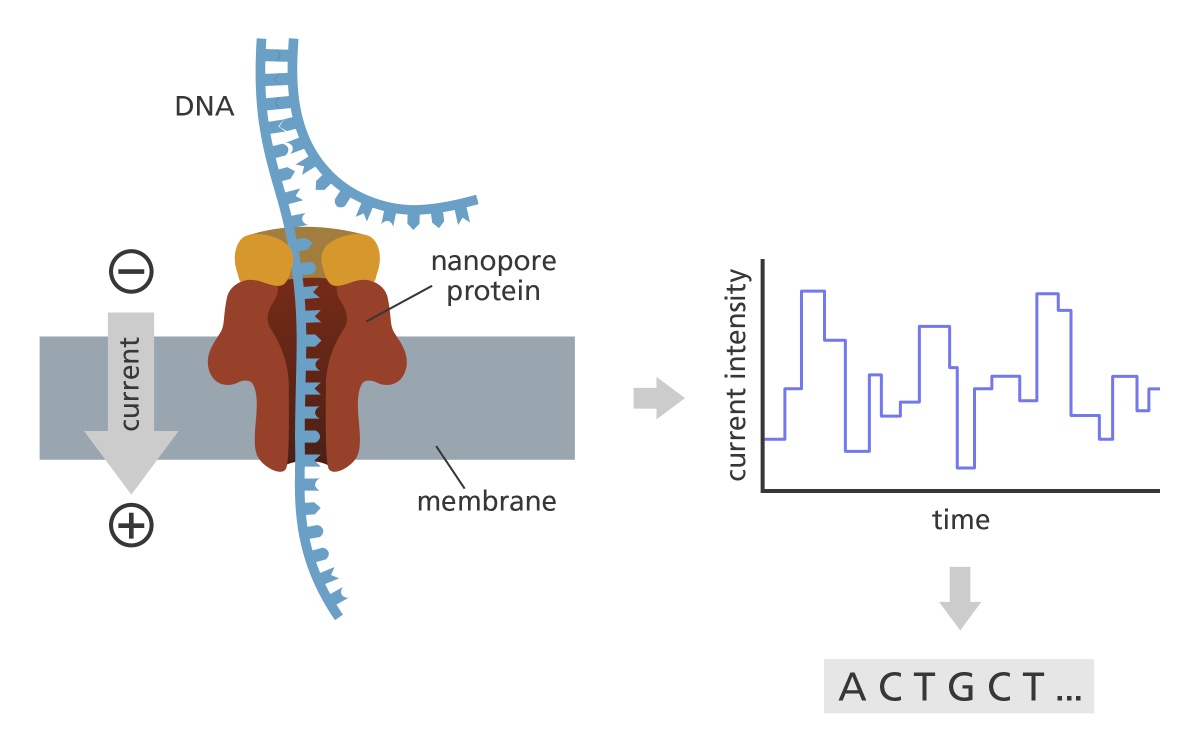
Overview of the PacBio sequencing strategy. (This image has been taken from https://www.yourgenome.org/wp-content/uploads/2022/04/ont-sequencing_yourgenome.png)#
As opposed to the previous technologies, this is a not a sequencing-by-synthesis approach. Instead, the (possibly very long) DNA-molecule is threaded through a small protein pore (alpha-hemolysin) that is embedded into an artificial membrane. A small motor-protein attached to the DNA molecule regulates the speed of the DNA traversal (approx 400 nt / sec). Once an electric potential is applied across the membrane the negatively charged DNA molecules will pass through the nanopore. Due to the low width of the nanopore (approx 1 nm) the DNA molecule just fits through and the different nucleotides currently within the flow cause a characteristic change of the otherwise constant electric current through the pore. The differences in the current are measured over time and can then be used to infer the sequence that passed through the pore.
High throughput is achieved through parallelization. In recent sequencers more than 2000 nanopores sequence in parallel per flow-cell. One further advantage is that the sequencing happens in real time. That is, only mere minutes after starting a run the first sequences can be collected, opening up a tremendous potential for clinical applications, where short turnaround times are critical.
Similar to PacBio sequencing also ONT sequencing works on DNA that has not been amplified. Just as the different nucleotides cause a characteristic change in the current, any modification of DNA, such as a methylation, will do the same. In general, the nanopore can record sequences of arbitrary length and instances of reads reaching a length of more than 1 Mb have been reported.
Key characteristics:
high throughput (up to 65 Gb / hour)
moderate to high error rate (1-15 %)
long read length (median of 20-30 kb)
very small hardware size (comparable to smartphone)
Further reading#
Further information on the different sequencing technologies: