Array-based genomics and transcriptomics#
Before high-throughput sequencing entered the stage, array-based measurement techniques were broadly used for interrogating the composition of sequencing samples in a qualitative and quantitative manner. Depending on the application, this technology is still being used today (e.g., for array-based genotyping).
The general principle of a micro-array is to expose an oligonucleotide with a specific sequence at a predetermined position. Arranged in a very dense grid, each specific location and sequence can then be addressed using a 2D coordinate. To increase signal strength and allow for quantitative readouts, each location (spot) contains multiple copies of the same oligonucleotide.
For measurement the array is then exposed to a library of short / fragmented target sequences that are fluorescence-labeled. Exploiting DNA base-complementarity, the target sequences specifically bind to the respective oligonucleotides and emit a measurable fluorescence signal.
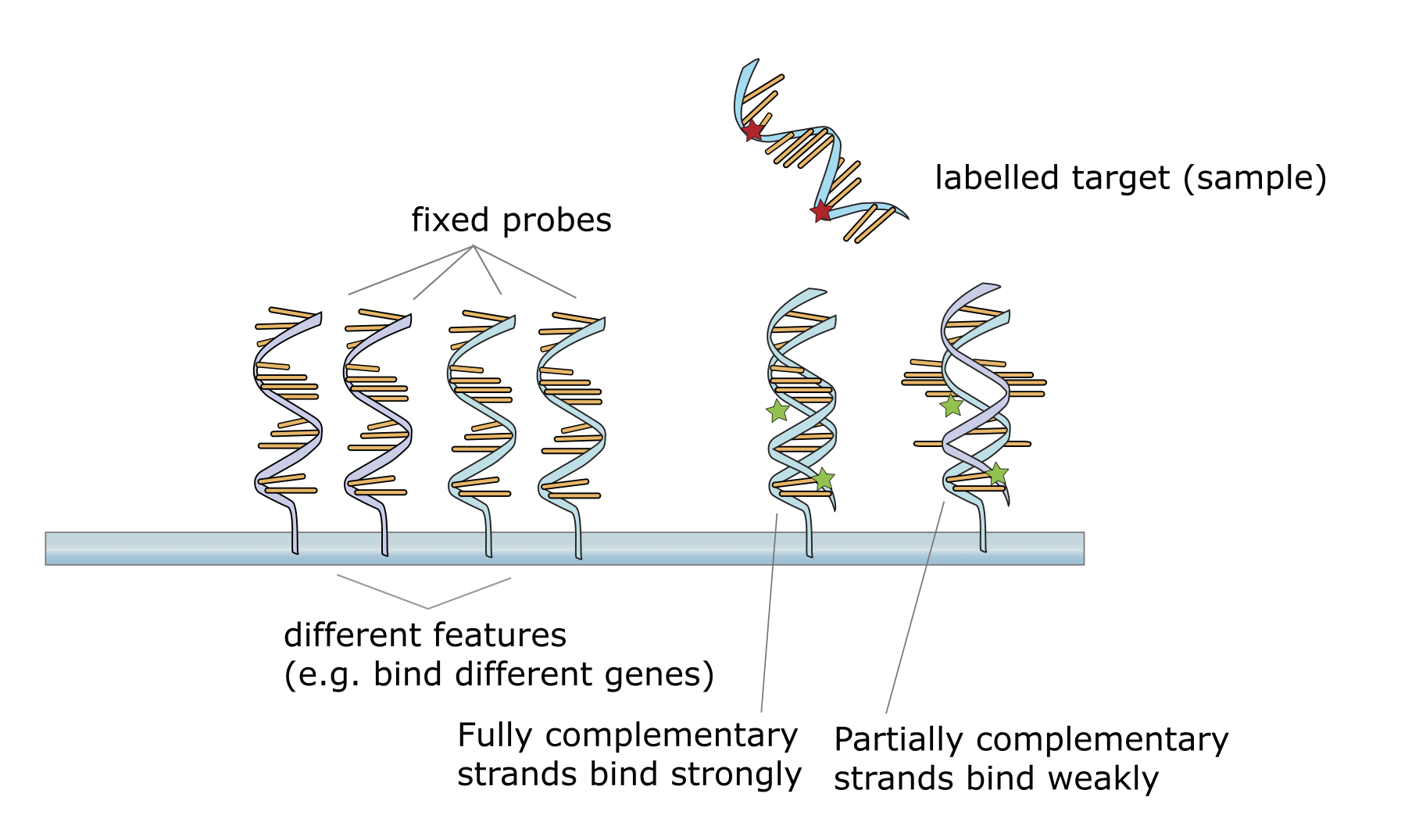
Schematic of the hybridization mechanism between fluorescence-labeled target and array oligo. (Source: https://en.wikipedia.org/wiki/DNA_microarray#/media/File:NA_hybrid.svg)#
Array-based measurements have found broad application in the past. A main motivation for the continued use in some areas is the cost-effectiveness. Also, a rich portfolio of analysis methods has been developed over the past years. Typical areas of application are SNP arrays, where each oligonucleotide probe encodes for a single nucleotide polymorphism together with its genomic context (to allow for highly specific detection); expression microarrays, where each oligonucleotide probe encodes a fraction of a gene or transcript that should be quantified; or exon arrays, where each oligonucleotide probe encodes an exon-exon junction and thus allows for measuring the presence / expression of a specific splice junction.
Despite their practical applications, arrays also show a number of limitations:
Restriction to known sequences - As all oligonucleotide sequences need to be specified during the design of the array, only known sequence features can be measured. Thus, the technology can not be used for the discovery of new features, such as novel exon-exon junctions or new sequence variants.
Limited dynamic range - As each spot only has a fixed (and limited) number of oligonucleotides that can bind to target sequences, all quantifications derived from array hybridizations have a saturation value that they cannot exceed. That is, it can happen that two very highly expressed genes both saturate the fluorescence signal and any relative difference in expression cannot be observed.
Measurement array - Like any other measurement technology, also arrays can produce erroneous measurements. Especially for highly expressed genes on a densely packed array, spill-over effects between neighboring spots can lead to false-positive expression measurements. Further, cross-hybridization and non-specific binding can create false-positive readouts.