Introduction to mOTUs#
mOTUs uses 10 universal marker genes to taxonomically profile metagenomes, to quantify metabolically active members in metatranscriptomics and to quantify differences between strain populations using single nucleotide variation (SNV) profiles.
Today we will show how to use the tool for the first and second of these tasks, profiling metagenomes and metatranscriptomes.
Running a single sample#
The most basic command is profile, which runs the entire mOTUs pipeline with default parameters.
submit motus profile -f <forward reads> -r <reverse reads> -o <output file> -t 12
We will have to put this command into a job submission script as described in the earlier session and then leave it to run in the queueing system.
Running individual steps#
Whilst this is running, let’s take a look at the individual steps of the pipeline:
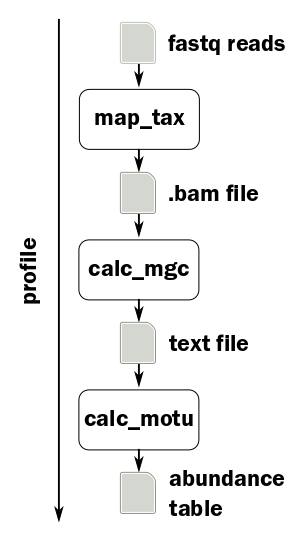
map_tax#
This command uses the program BWA to align the reads against the mOTUs database. The output is a SAM or BAM file (Sequence or Binary Alignment/Map), which is a standard format used by the SAMtools set of utilities.
submit motus map_tax -f <forward reads> -r <reverse reads> -o <bam file> -b -t 12
calc_mgc#
The intermediate step calculates the number of reads aligned to each marker gene cluster, weighted by gene length.
submit motus calc_mgc -n <sample name> -i <bam file> -o <mgc file>
The output format consists of two columns: the name of the marker gene cluster and the count.
calc_motu#
The final step controls the output format of the final abundance table.
submit motus calc_motu -n <sample name> -i <mgc file> -o <output file>
mOTUs output#
The standard output format consists of two columns: the mOTU name and the relative abundance of the mOTU in the sample. Note that the table lists all possible mOTUs, i.e.: it includes zeros.
#consensus_taxonomy M01.1-V1-stool-metaG
Kandleria vitulina [ref_mOTU_v2_0001] 0.0000000000
Methyloversatilis universalis [ref_mOTU_v2_0002] 0.0000000000
Megasphaera genomosp. [ref_mOTU_v2_0003] 0.0000000000
Streptococcus anginosus [ref_mOTU_v2_0004] 0.0000000000
Streptococcus anginosus [ref_mOTU_v2_0005] 0.0000000000
...
unknown Clostridiales [meta_mOTU_v2_7795] 0.0000898639
unknown Bacteroidetes [meta_mOTU_v2_7796] 0.0000000000
unknown Opitutae [meta_mOTU_v2_7797] 0.0000000000
unknown Roseburia [meta_mOTU_v2_7798] 0.0000000000
unknown Firmicutes [meta_mOTU_v2_7799] 0.0000000000
unknown Clostridiales [meta_mOTU_v2_7800] 0.0000000000
-1 0.0248427818
Merging output#
When you have run mOTUs on multiple samples, you can combine the output into a single, tab-separated table with the merge command.
motus merge -i <motu file 1>,<motu file 2>,... -o <output file>
motus merge -d <directory> -o <output file>
Since you have been given metagenomic and metatranscriptomic data for the same sample, run both with default parameters and merge the two sets of results into one .motu file.