Introduction to Unix 1¶
General information¶
Topic overview¶
In this lesson you will learn about Unix: an operating system that runs on almost all high performance computing (HPC) servers, which we interface with via the command line.
Learning objectives¶
Learning Objective 1 - You understand the inner workings of the computer: how files are organized and how processes are run
You execute commands on the command line
You navigate the file system using commands
You create, modify, move, copy files and directories
Learning Objective 2 - You recognize the difference between local and remote servers and are able to communicate within and between them
You transfer files back and forth between your local computer and the remote server
What is Unix?¶
Unix is a family of operating systems - sets of programs that manage the use of computer hardware resources by software applications. It thereby allows multiple users to access a computer simultaneously, perform parallelized computations, exchange data and share hardware resources. This diagram shows an oversimplified view of an operating system - the kernel (in blue) is a core program that is always active and controls interactions between the hardware (in green), and software (in red).
Technically, our servers use a version of Linux, a Unix-like operating system. Furthermore, even major operating systems such as Android and MacOS are Unix-based underneath.
The command¶
Commands are our tool to tell the computer what to do. Most commands have options and arguments. Arguments are often essential for a command to operate properly; they are the pieces of information required by a command, such as a file name. Options are, of course, optional, and offer ways to modify the way the command works.
For instance, echo will take any text you give it as an argument and then send it back to you as output:
# My first command
echo 'Hello World!'
If you use the option -n, then it will not add a ‘new line’ to the end of the output:
# My second command
echo -n 'Hello World!'
Some commands end up with very complex structures, because they can have many options and arguments. In general, options will be of the format -a where a is a single letter or --word where word is a string (a series of letters, in computer terms).
Note: the command line is case-sensitive! So it does matter if you write -a or -A.
Getting help¶
The man command will show a manual for most basic commands, providing the correct syntax to use it and the various options available.
# Read the manual
man ls
Other programs have different ways to provide help on how to use them. An online tutorial is best, or a comprehensive manual, but sometimes you only have the command line to help you.
# Help please!
python3 -h
python3 --help
Useful command line tricks¶
You can use the up and down arrow keys to navigate through previously used commands (known as your history) and repeat or modify them.
Windows: To copy text from the terminal you will have to highlight it and right-click to use the in-browser menu and copy. Similarly you have to use the in-browser menu to paste into the terminal. The reason for this is that Ctrl + c and Ctrl + v have effects inside the terminal.
Mac: You can fortunately use Cmd + c and Cmd + v to copy and paste as normal. You can use Ctrl and various keys for in-terminal commands.
When typing a command or file name, you can press the ‘tab’ key to auto complete what you are typing. If there are multiple commands or files with similar names, auto completion will fill in as far as the first ambiguous character before you have to give it some more input. This method makes it much less likely that you make a spelling error. Also, if you double press the ‘tab’ key all the available options to complete will be shown.
Pressing Ctrl + c will send an interrupt signal that cancels the currently running command and brings you back to the command line.
Pressing Ctrl + r will allow you to search through your command history.
Pressing Ctrl + l will clear the screen.
See previous commands by typing history and pressing enter.
Double-click to select a word, triple-click to select a line
Using a # character allows you to make comments that have no effect when run.
Exercise 1.1
Try to echo “My first command”
#echoing "My first command"
echo 'My first command'
Use the arrow key to execute the same command again
# Press the up arrow once and the last command appears
echo 'My first command'
Try typing e then pressing tab twice, what do you see?
# You see all the possible commands that start with "e" when you press tab twice after entering “e”
e2freefrag edquota era_check eu-readelf
e2fsck efibootdump era_dump eu-size
e2image efibootmgr era_invalidate eu-stack
e2label efikeygen era_restore eu-strings
e2mmpstatus efisiglist esac eu-strip
e2undo efivar escputil eutp
e4crypt egrep espdiff eu-unstrip
e4defrag eject espeak-ng eval
eapol_test elfedit ether-wake evince
easy_install-2 elif ethtool evince-previewer
easy_install-2.7 else eu-addr2line evince-thumbnailer
easy_install-3 enable eu-ar evmctl
easy_install-3.6 encguess eu-elfclassify ex
ebtables enchant-2 eu-elfcmp exec
ebtables-restore enchant-lsmod-2 eu-elfcompress exempi
ebtables-save enscript eu-elflint exit
echo env eu-findtextrel exiv2
ed envsubst eu-make-debug-archive expand
edgepaint eog eu-nm export
edid-decode eps2eps eu-objdump exportfs
editdiff eqn eu-ranlib expr
Try adding c to make ec and pressing tab again. What happens?
# The command autocompletes after adding the “c” to the “e”
echo
Try to copy/paste your echo command “echo ‘My first command’”
# Note that ctrl + c to copy does not work in windows terminal or mobaxterm - instead highlighting text automatically copies it
# In mobaxterm ctrl + v to paste also does not work - instead right-click to paste
echo 'My first command'
Try to clear the screen, can you still paste your echo command?
# To clear the screen use ctrl + l or the command 'clear' and you can still paste the command
echo 'My first command'
Try to echo ‘My first command ‘once with the -n option and once with the -N option. What do you notice?
# echo -n does not add a new line to the output
echo -n 'My first command'
My first command[]$
# The -N option does not exist therefore “echo” will ill interpret '-N' as characters to display
echo -N 'My first command'
-N My first command
Try to look at the manual of the echo command.
# man gives you the manual of the function
man echo
The file system¶
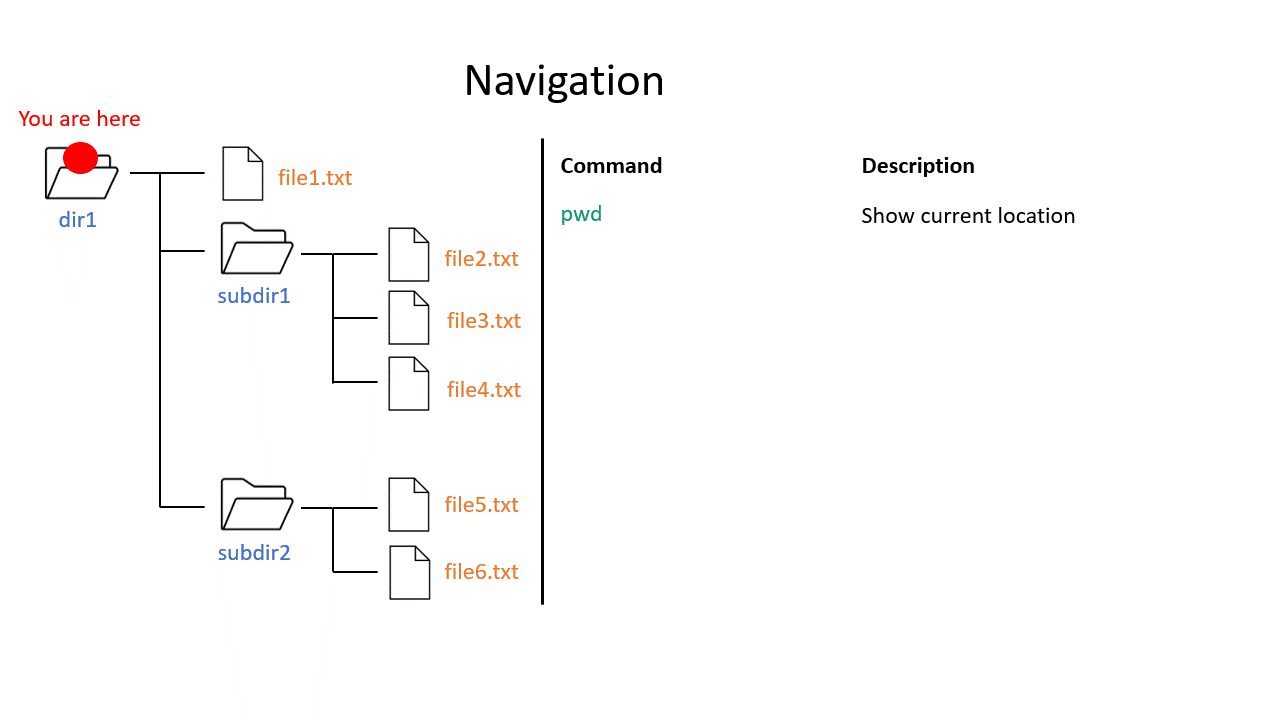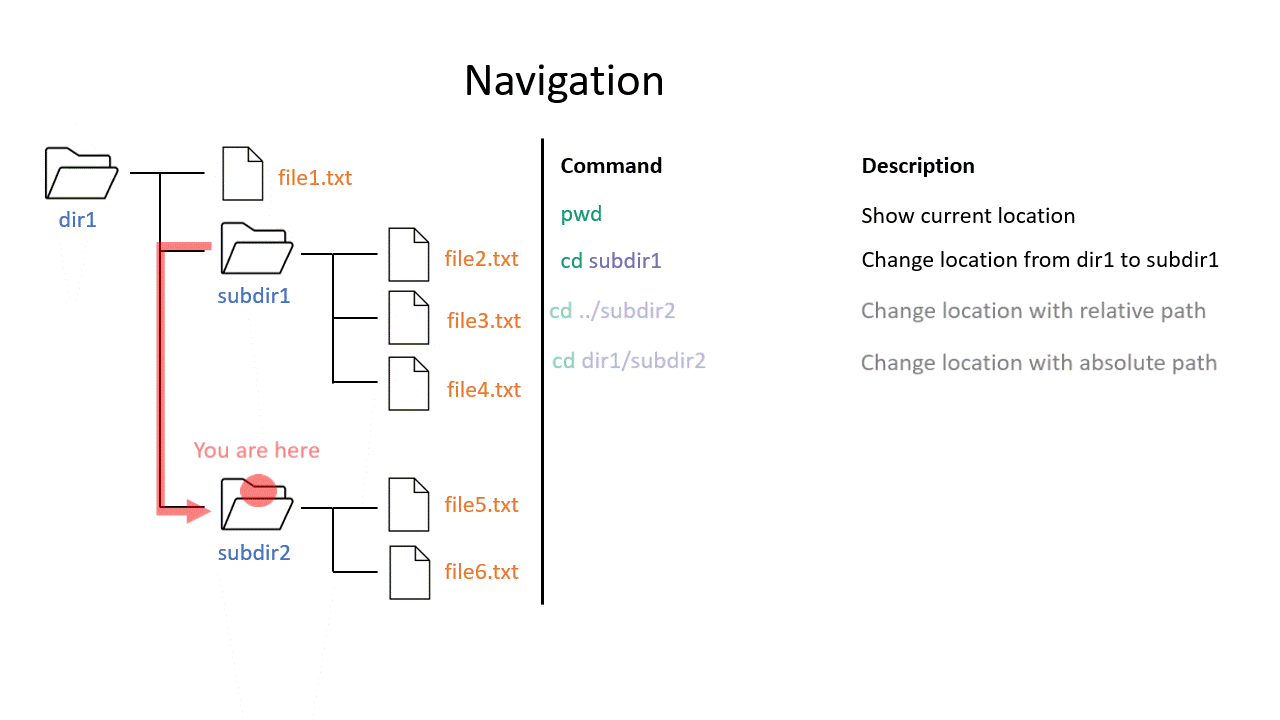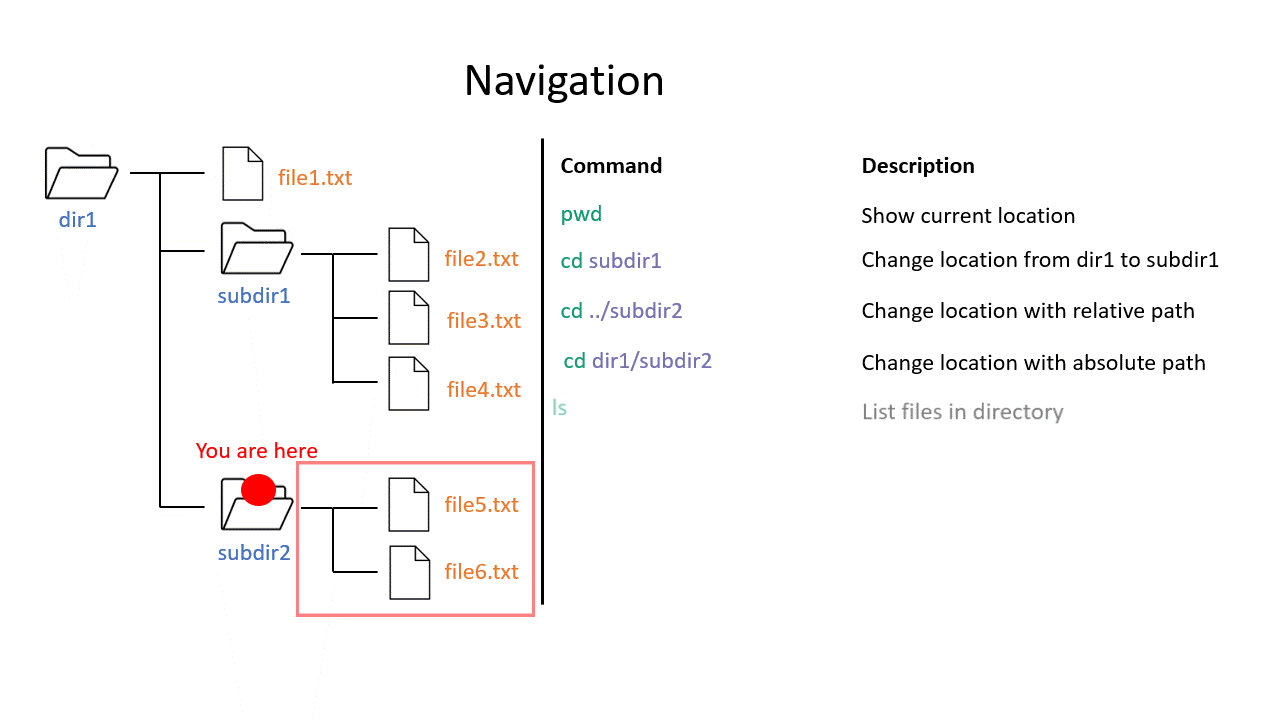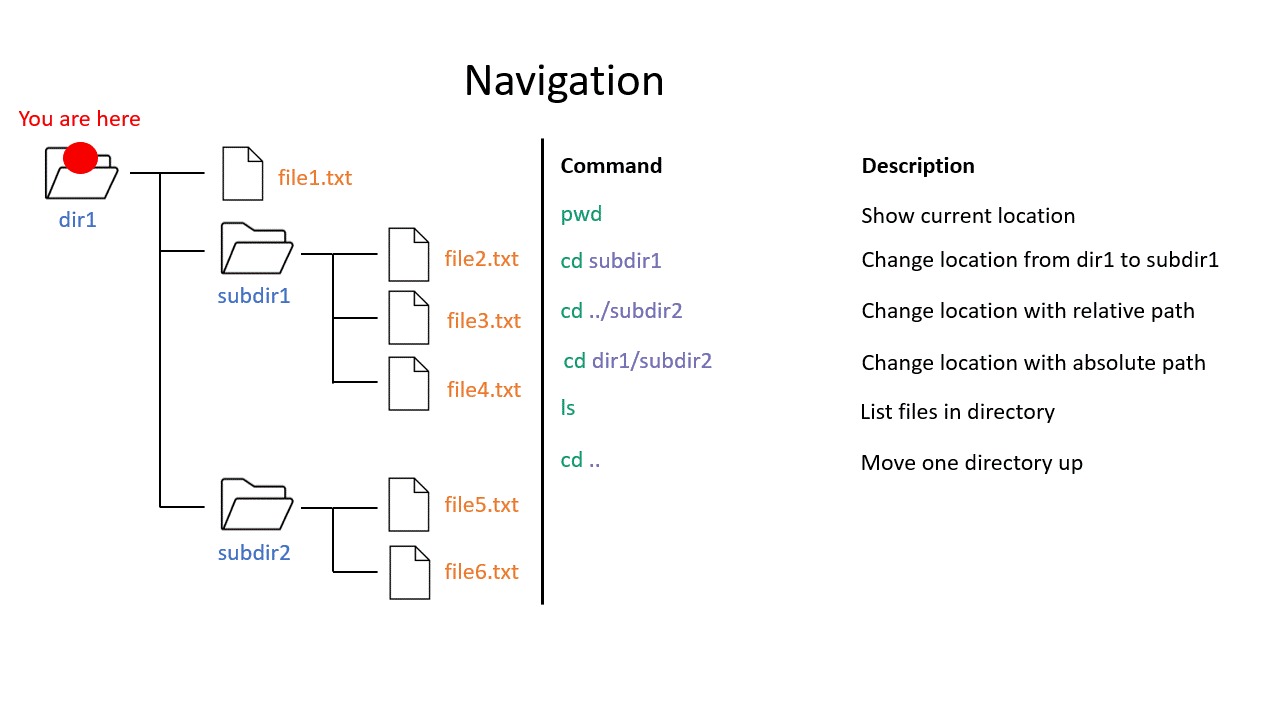
You may be used to the file system in Windows or Mac OS X, where directories (or folders if you prefer) can contain files and more directories. The Unix filesystem is structured in the same way - as a tree - that begins at the ‘root’ directory ‘/’. Directories are separated by slash characters /.
Click on the image below to see what your file system could look like. In this example the root directory is dir1.
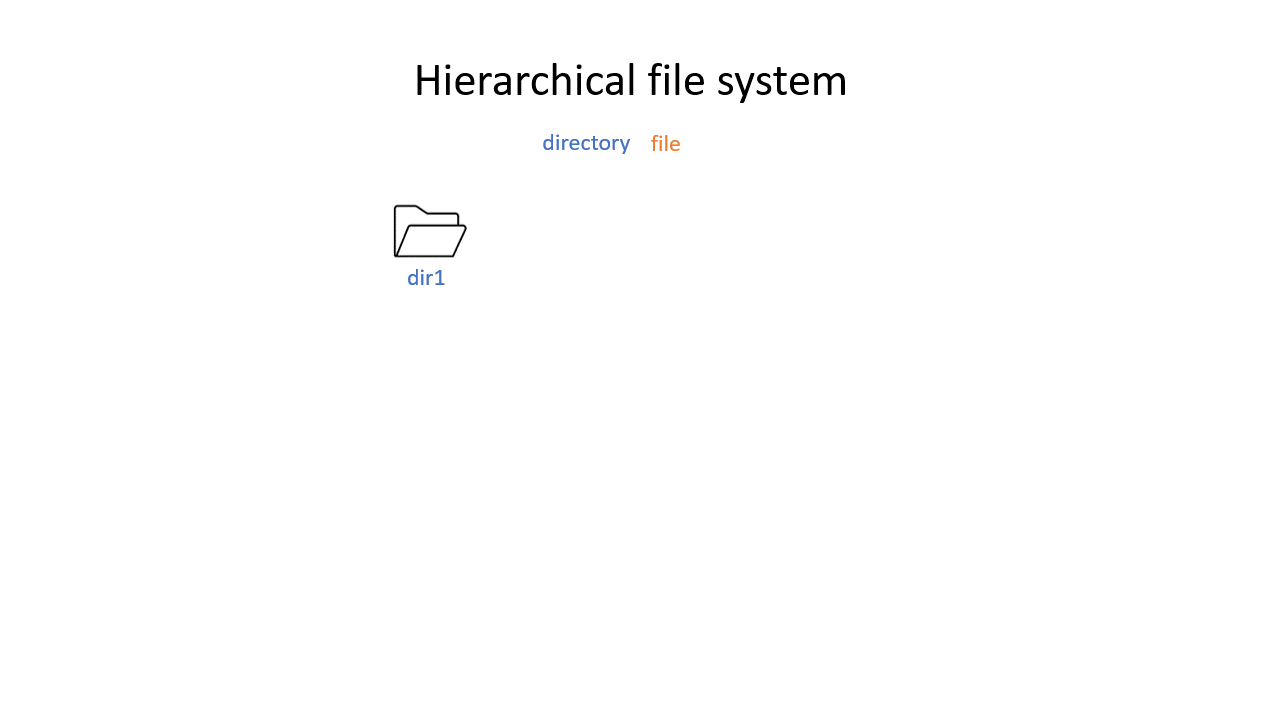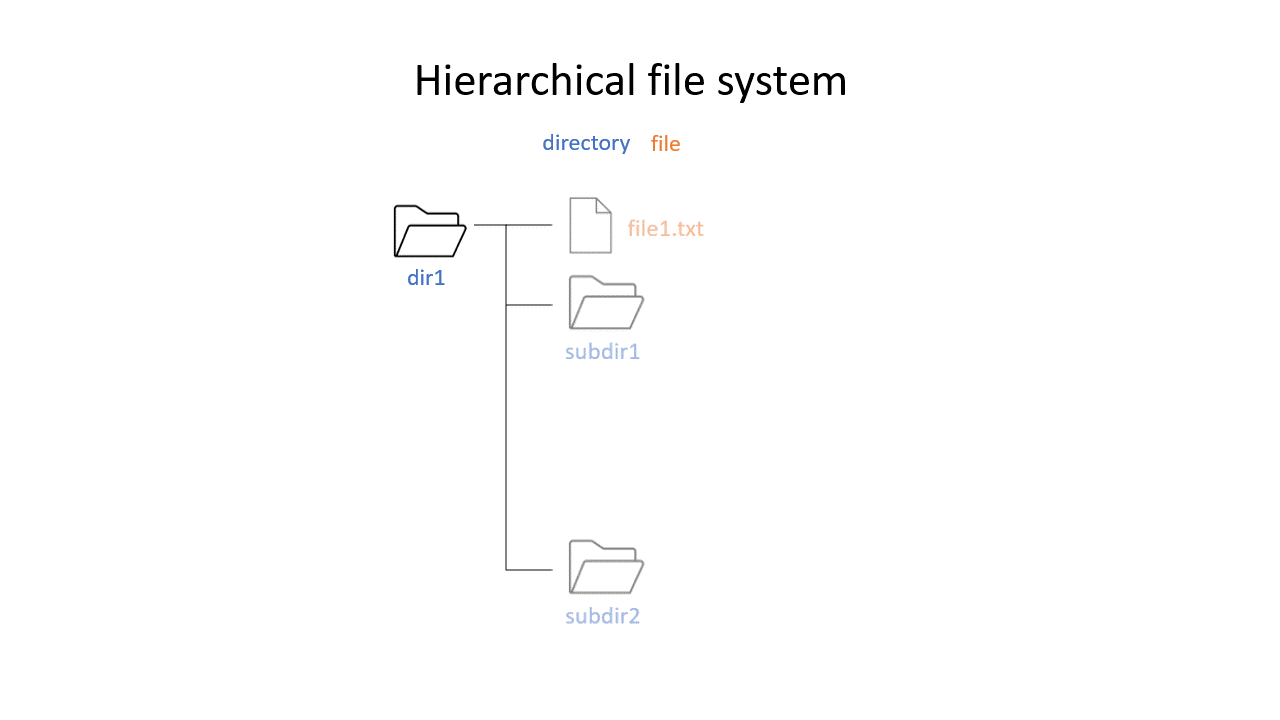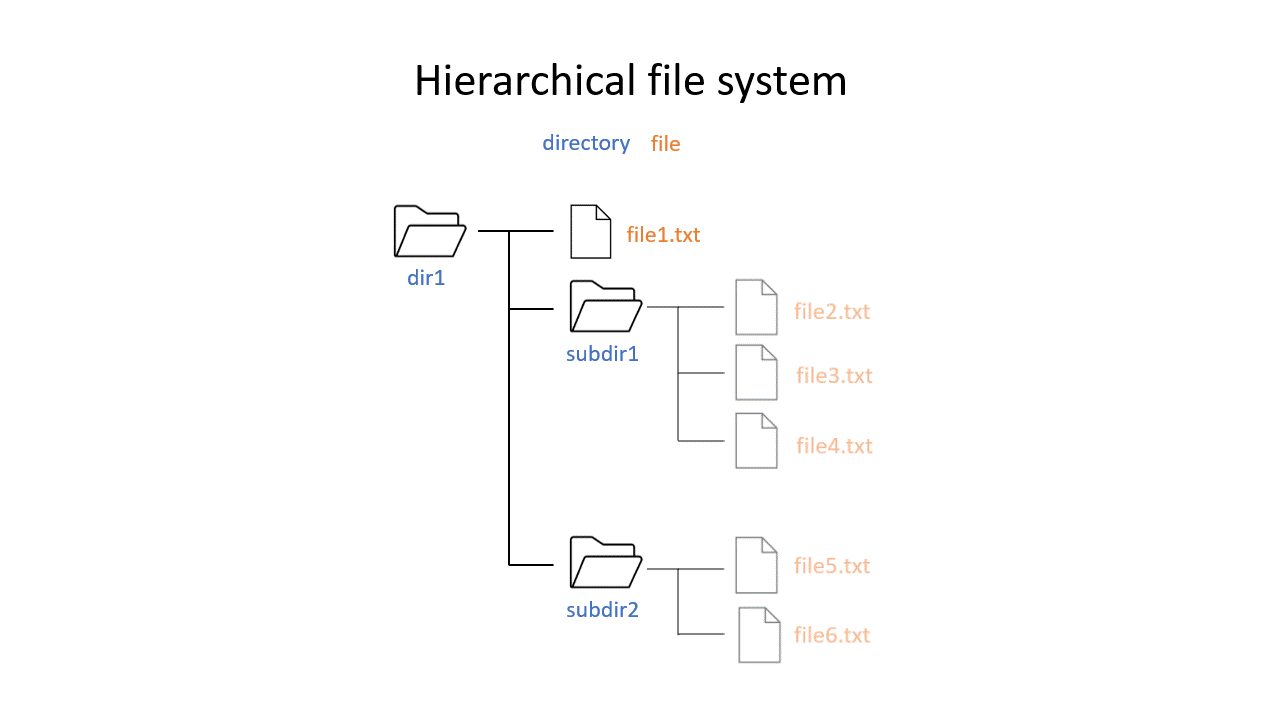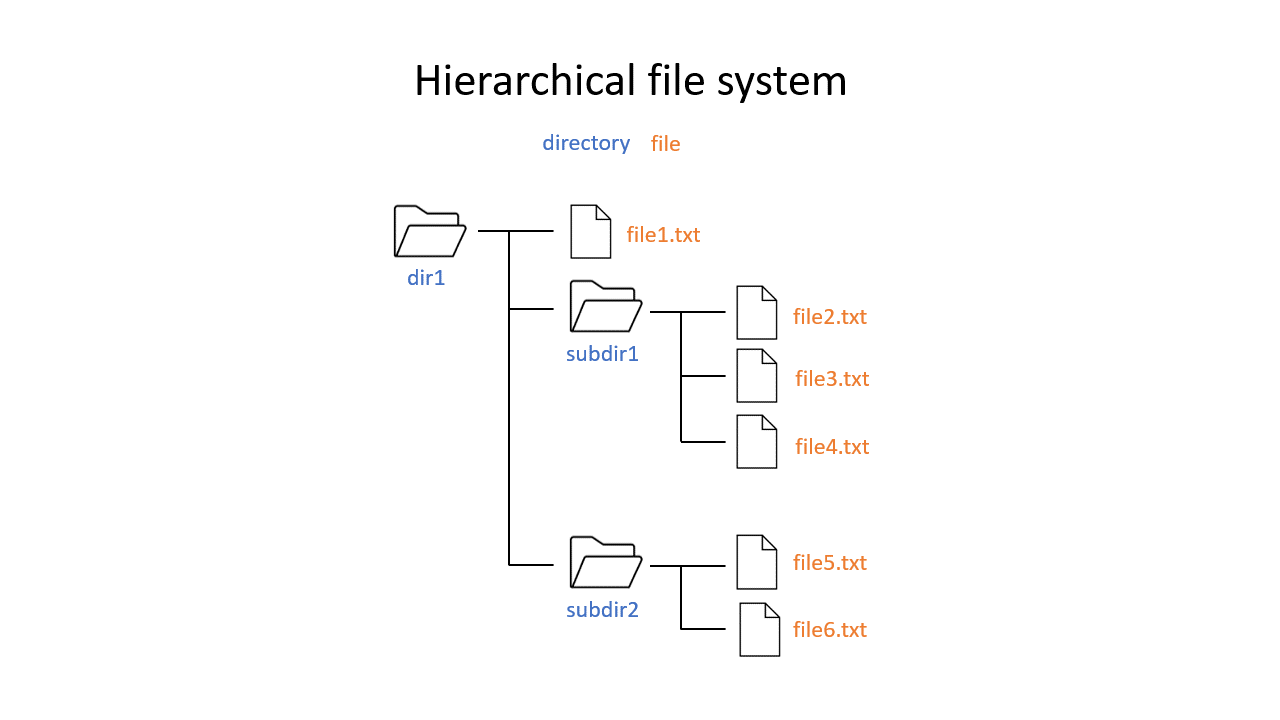
When you work on the command line, you are located in a directory somewhere in this tree. There are two ways to refer to a location: its absolute path, starting at the root directory, or its relative path, starting in the current directory.
# Absolute path
/nfs/course/home/<user_name>
# Relative path
../../home/<user_name>
The .. refers to the directory above a location, so the relative path here goes up twice, then back down to your home directory. If a path starts with ~/ then it refers to your home directory. If a path starts with ./ then it refers to the current directory.
# References the level above
../
# References the home directory
~/
# References the current directory
./
Wildcards¶
When providing a file path as an argument to a command, it is often possible to provide multiple file paths using wildcards. These are special characters or strings that can be substituted for a matching pattern. For many commands using wildcards allows you to execute the associated action on each file that matches the pattern, though this obviously does not work in all cases.
? matches any single character
* matches any number of any characters
[…] matches any character within the brackets
{word1,word2,…} matches any string inside the brackets
For instance:
# Pattern matching
ls /nfs/teaching/551-0132-00L/1_Unix1/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/* # lists all files in the ecoli directory
ls /nfs/teaching/551-0132-00L/1_Unix1/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/*.fna # lists all nucleotide fasta files there
ls /nfs/teaching/551-0132-00L/1_Unix1/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/*.f?a # lists all nucleotide and protein fasta files there
Basic file operations¶
cp copies a file from one location to another. The example will copy a file containing the genome sequence of E. coli K12 MG1655 to your home directory.
# Copy
cp <source> <destination>
cp /nfs/teaching/551-0132-00L/1_Unix1/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna ~/
mv moves a file from one location to another. The example actually renames the file, because the destination is not a directory. Thus you can move and rename a file with the same command.
# Move or rename
mv <source> <destination>
mv ~/GCF_000005845.2_ASM584v2_genomic.fna ~/E.coli_K12_MG1655.fna
rm removes a file, so use it with care.
# Remove
rm <path_to_file>
rm ~/E.coli_K12_MG1655.fna
mkdir creates a new directory with the given name.
# Make directory
mkdir <path to directory>
mkdir genomes
rmdir removes an empty directory.
# Remove an empty directory
rmdir <path to directory>
rmdir genomes
Click on the image below to see what every command will do within your file system.
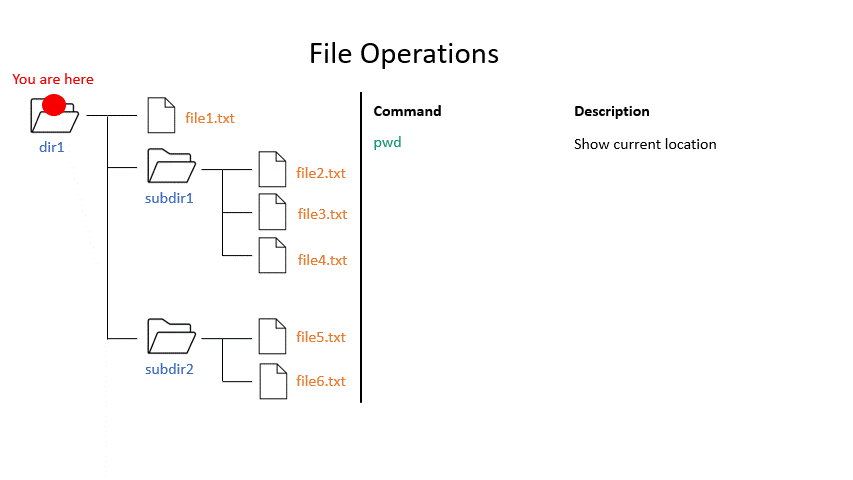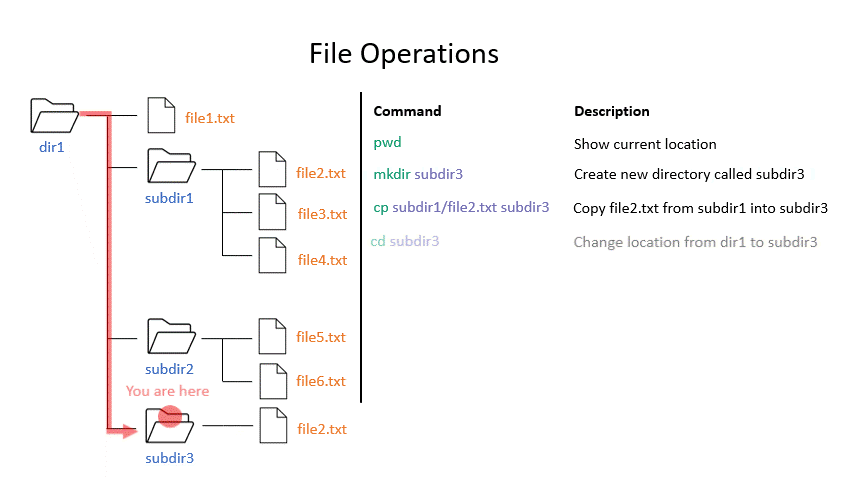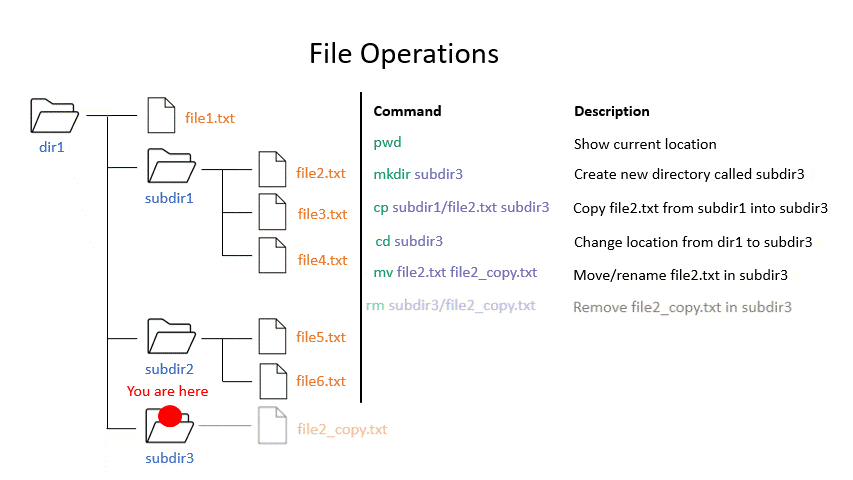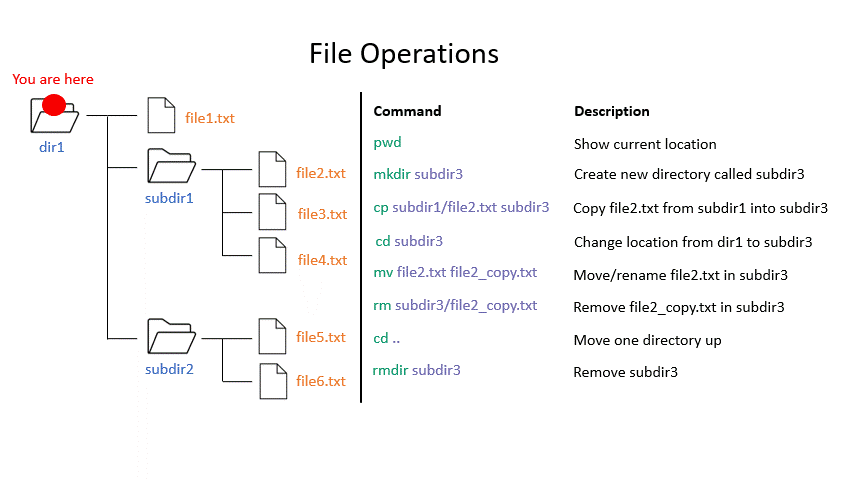
Exercise 1.3
Create three new directories called “genomes”, “test” and “in_class” in your home folder
# First go to your home folder
cd
# Use the mkdir function to create a directory
mkdir genomes
mkdir test
mkdir in_class
Delete the test directory
# Use the rmdir function to remove a directory
rmdir test
Copy the
/nfs/teaching/551-0132-00L/1_Unix1/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fnafile into your new directory “genomes”
# Use the cp function to copy. cp <source> <destination>
cp /nfs/teaching/551-0132-00L/1_Unix1/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna ~/genomes
Rename the file to “E.coli_K12_MG1655.fna”
# Use the move function to rename a file mv <source> <destination>
# Enter the genomes directory
cd genomes
# Rename file
mv GCF_000005845.2_ASM584v2_genomic.fna E.coli_K12_MG1655.fna
Use the help option of the ls function to find out which option will give file size in a human-readable format. Remember that ls -l will give a table of file information including size.
# ls -l will give you a table of information but the file sizes are long and hard to read
ls -l
# ls --help lists all the options possible
ls --help
# The -h option makes file sizes human-readable, however file sizes are only printed when you use the -l flag so you must use both
ls -l -h
ls -lh
# The size should be 4.5M
Using man and cp, find out how to copy a directory.
# Enter home directory
cd
# Create two directory
mkdir dir1
mkdir dir2
# Try to copy dir1 into dir2
cp dir1 dir2
cp: dir1 is a directory (not copied).
# If you check 'man cp', you see that you have to use -R:
man cp
cp -R dir1 dir2
# Check if the directory has been copied
ls dir2
File name conventions¶
In Unix systems there are only really two types of files: text or binary. The file name ending (.txt or .jpg) doesn’t really matter like it does in Windows or Mac OS, however it is used to indicate the file type by convention. Some file types you will encounter include:
.txt - A generic text file.
.csv - A ‘comma separated values’ file, which is usually a table of data with each line a row and each column separated by a comma.
.tsv - A ‘tab separated values’ file, which is the same but separated by tab characters.
.fasta or .fa - A fasta formatted sequence file, in which each sequence has a header line starting with ‘>’.
.fna - A fasta formatted nucleotide sequence file, usually gene sequences.
.faa - A fasta formatted protein sequence file.
.sh - A ‘shell script’, which contains commands to run.
.r - An R script, which contains R commands to run.
.py - A python script, which contains python commands to run.
.gz or .tar.gz - A file that has been compressed using a protocol called ‘gzip’ so that it takes up less space on the disk and transfers over the internet faster.
Other useful file operations¶
Transferring files between computers¶
There are many different protocols for transferring files between computers. You may have heard of FTP - File Transfer Protocol - which is a non-secure but commonly used example. A more secure file transfer protocol is SCP - Secure Copy Protocol, and programs such as WinSCP use it. The command scp is an easy way to transfer a file immediately between the server you are working on and another (or two different servers!). Another command to copy files is rsync, which can be used with many options such as preserving the ownership and date of creation of a file (and much more).
Be aware that all the scp commands shown below have to be executed on your local computer. Thus if you are currently connected to a server, you first have to exit this connection to be connected to your local computer again (Windows Command or Mac Terminal).
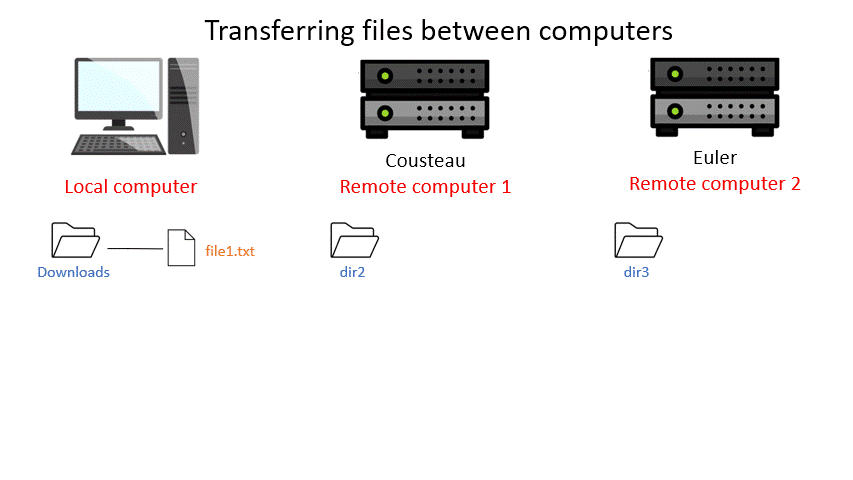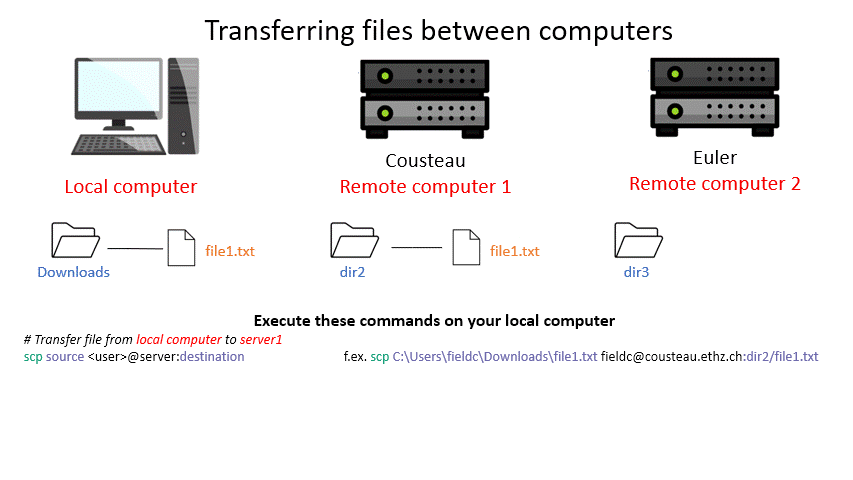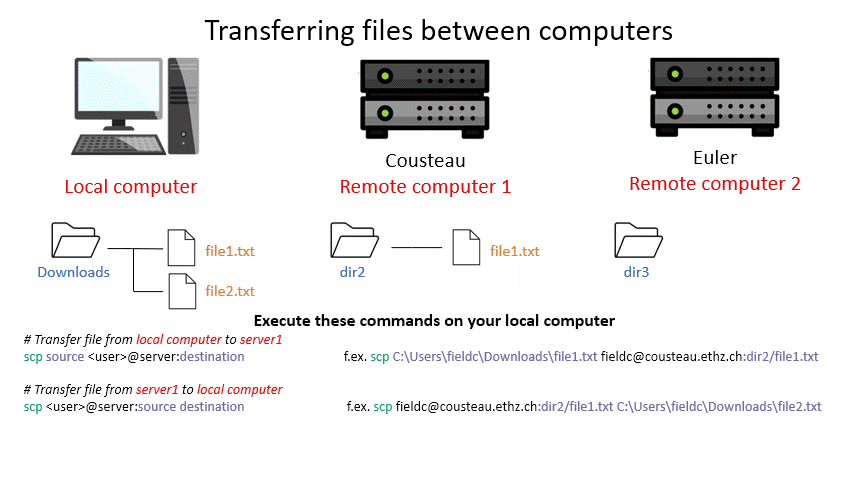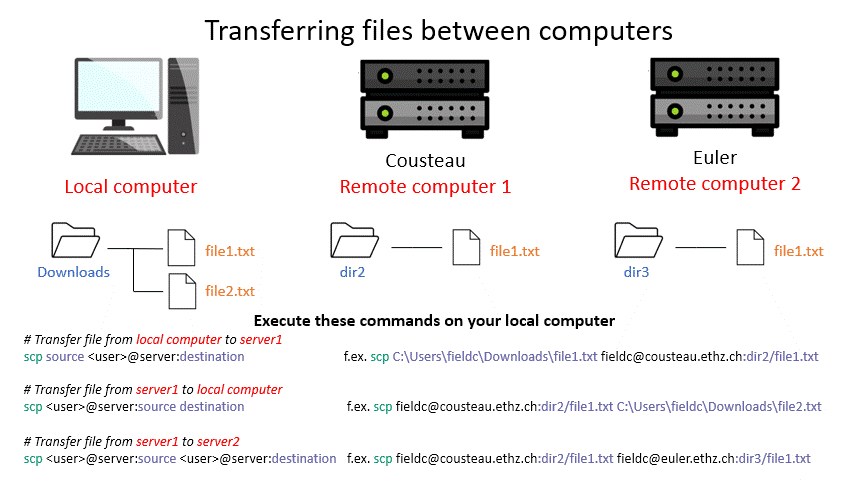
Tip: Remember that with the ‘tab ‘ key you can auto complete and see the available options by double pressing. This can make finding a file you want to upload way easier. (Note: This works only for the machine you are currently on.)
# Secure CoPy
man scp
scp /path/to/source user@server:/path/to/destination # local server (where the command is executed) to remote server
scp <user>@server:/path/to/source /path/to/destination # remote server to local
scp <user>@server1:/path/to/source <user>@server2:/path/to/destination # remote server 1 to remote server 2
# Download an E. coli genome from the server to your local computer
# First open Windows Command or Mac Terminal on your local computer
scp <user>@cousteau.ethz.ch:/nfs/teaching/551-0132-00L/1_Unix1/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna .
# Note that the local path is just "." - this means "here", i.e.: your working directory
# Copy the E.coli genome (or any file) from your local computer to the home folder on the server
# Again, on your local system, run the following commands in Windows Command or Mac Terminal
scp GCF_000005845.2_ASM584v2_genomic.fna <user>@cousteau.ethz.ch:~/
# Copy the E.coli genome from cousteau to euler, executed on cousteau
scp GCF_000005845.2_ASM584v2_genomic.fna <user>@euler.eth.ch:~/
# Copy the E.coli genome from cousteau to euler, executed on your own computer
scp <user>@cousteau.ethz.ch:GCF_000005845.2_ASM584v2_genomic.fna <user>@euler.ethz.ch:~/
Warning
Windows Paths
Windows terminal is not unix-like - instead it is based on MS-DOS and uses some different commands and conventions. Paths on Windows terminal use backslashes instead of forward slashes, and the root is the letter assigned to the drive where the path is. For instance a file in my downloads folder might have the path:
C:\Users\fieldc\Downloads\file.txt
Sometimes you want to download a file directly from the internet to the server, rather than going via your local machine. For using this command, you have to connect to the server first and then wget allows you to download files in this way.
# Download from the internet
wget source-URL
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/482/265/GCF_000482265.1_EC_K12_MG1655_Broad_SNP/GCF_000482265.1_EC_K12_MG1655_Broad_SNP_genomic.fna.gz
Compressing and decompressing files¶
Files can be compressed to take up less space on the hard drive (disk), or for transfer over the internet. The file you downloaded is an example, and we can decompress it using the gunzip command:
# Decompress a file
gunzip GCF_000482265.1_EC_K12_MG1655_Broad_SNP_genomic.fna.gz
If you ever need to compress a file, for instance to send it to someone, you can use the gzip command:
# Compress a file
gzip GCF_000482265.1_EC_K12_MG1655_Broad_SNP_genomic.fna
Exercise 1.4
Download the mystery image
/nfs/teaching/551-0132-00L/1_Unix1/mystery_image.jpgfrom the server to your local machine. What is it?
On the server, download the E. coli file from the example above to your home folder.
# Make sure I am in my home directory
cd ~
# Download the file
wget https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/482/265/GCF_000482265.1_EC_K12_MG1655_Broad_SNP/GCF_000482265.1_EC_K12_MG1655_Broad_SNP_genomic.fna.gz
Decompress the file.
#Decompress it
gunzip GCF_000482265.1_EC_K12_MG1655_Broad_SNP_genomic.fna.gz
Working with files¶
Looking at files¶
The command cat displays the entire contents of a file directly on the terminal. For large files this can be disastrous, so remember that you can cancel commands in progress with ctrl + c.
# ConCATenate
cat E.coli_K12_MG1655.fna
The command head displays only the first 10 lines of a file directly on the terminal. If you look at the available options for the command, -n x outputs the first x lines instead, and using a negative number outputs the lines except for the last x.
# Show file head
head E.coli_K12_MG1655.fna
head -n 1 E.coli_K12_MG1655.fna
The command tail displays only the last 10 lines of a file directly on the terminal. It has similar options to head; -n x outputs the last x lines, and using a positive number +x (note the “+” character) outputs the lines except for the first x.
# Show file tail
tail E.coli_K12_MG1655.fna
The command less is a versatile way to look at a file in the command line. Instead of showing you the contents of a file directly on the terminal, it ‘opens’ the file to browse. You can use the arrow keys, page up, page down, home, end and the spacebar to navigate the file. Pressing q will quit. A number of useful options exist for the command, such as showing line numbers or displaying without line wrapping. It also has a search feature that we will cover later.
# Browse file
less E.coli_K12_MG1655.fna
The command wc is a command that will quickly count the number of lines, words and characters in a file, including invisible characters like ‘newline’ and whitespace. Its options allow you to specify which value to return, otherwise it gives all three.
# Count things
wc E.coli_K12_MG1655.fna
Exercise 1.5
Use cat to look at the E. coli genome file you copied last time, is it suitable for looking at this file?
# Your file should be located in your genomes directory
cd ~/genomes/
# If you have not copied and renamed the file yet, you can use these commands to do so
cp /nfs/teaching/551-0132-00L/1_Unix1/genomes/bacteria/escherichia/GCF_000005845.2_ASM584v2/GCF_000005845.2_ASM584v2_genomic.fna ~/genomes
cd ~/genomes
mv GCF_000005845.2_ASM584v2_genomic.fna E.coli_K12_MG1655.fna
# Looking at the file
cat E.coli_K12_MG1655.fna
# Press ctrl + c to cancel the command
Use head and tail to examine the first and last 10 lines of the genome file. Now try to look at the first and last 20 lines.
# Look at the first 10 lines (10 is the default value)
head E.coli_K12_MG1655.fna
# Look at the last 10 lines
tail E.coli_K12_MG1655.fna
# Look at the first 20 lines
head -n 20 E.coli_K12_MG1655.fna
# Look at the last 20 lines
tail -n 20 E.coli_K12_MG1655.fna
Use less to look at the genome file. Navigate through the file with the keys listed above, then return to the Terminal.
# Looking at the genome file
less E.coli_K12_MG1655.fna
#press q to quit
Use the man command we learned to read about the wc command.
# Read about the wc command
man wc
Can you find out how many lines are in the genome file with the wc command?
# Count the number of lines in the file
wc -l E.coli_K12_MG1655.fna