Session 1.2. Setup and Linux Introduction¶
Infrastructure¶
The resources for this course are provided in the form of RStudio Server running on a designated ETH machine with sufficient storage and computational capacity for all data analyses. The data you create will be provided to you after the course via our website.
Connection to RStudio Server¶
Each of you should be able to log in into the RStudio Server with your ETH credentials with a browser.
Once you access the RStudio server you’ll see a graphical user interface similar to a local version of RStudio.
Apart from the RStudio console you’ll find, next to it, the “Terminal” through which you can interact with the server using Linux commands. We will shortly practice some basic commands in the following tutorial.
Linux Introduction¶
Research in biology relies increasingly on computational analyses, but state-of-the-art tools often lack a graphical user interface or web service. Use of the command line is essential for such tools, but also offers increased power and flexibility for others.
In order to go through theis Block Course we have to know some basic commands for:
Locate files and folders
Move to specific locations
Inspect, edit, move, remove or copy files and folders
The structure of a command¶
Most commands have options and arguments. Arguments are often essential for a command to operate properly; they are the pieces of information required by a command, such as a file name. Options are, of course, optional, and offer ways to modify the way the command works.

For instance, echo will take any text you give it as an argument and then send it back to you as output:
# My first command
echo 'Hello World!'
If you use the optional flag -n then it will not add a ‘new line’ to the end of the output:
# My second command
echo -n 'Hello World!'
Some commands end up with very complex structures because they can have many options and arguments. In general, options will be of the format ‘-a’ where a is a single letter or ‘–word’ where word is a string (a series of letters, in computer terms).
Note: the command line is case-sensitive!
Useful tricks¶
You can use the up and down arrow keys to navigate through previously used commands and repeat or modify them.
By default in MobaXterm, you have to right-click the mouse to use the copy and paste commands, however this can be changed to the more common ‘highlight-to-copy’ method employed on most linux systems, where right-click will paste whatever is in the clipboard.
When typing a command or file name, you can press the ‘tab’ key to autocomplete what you are typing. However, if there are multiple files with similar names, it may only fill in as far as the first ambiguous character before you have to give it some more input. This method makes it much less likely that you make a spelling error.
If a command stops responding (perhaps it has crashed) then pressing Ctrl+C will send an interrupt signal that often cancels the command and brings you back to the command line.
The File System¶
You are probably used to the file system in Windows or MacOS, where directories can contain files and more directories. The linux filesystem is structured in the same way, as a tree, that begins at the ‘root’ directory /.
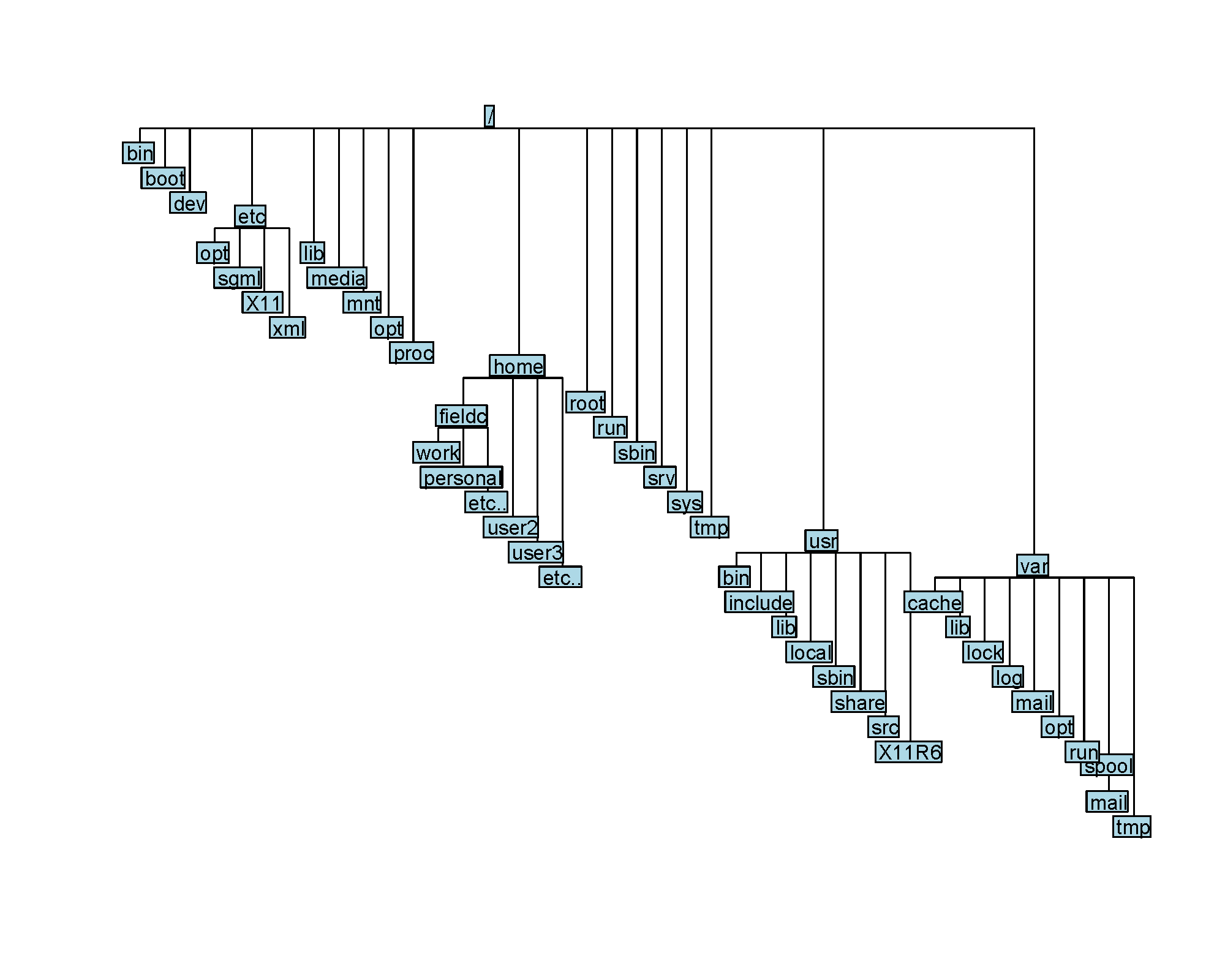
When you are working in the command line, you are located in a directory somewhere in this tree. There are two ways to refer to a location: its absolute path or its relative path.
DO NOT RUN THE CODE BELOW
# Absolute path
/nfs/course/home/guillems
# Relative path
../../home/guillems
The .. refers to the directory above a location, so the relative path here goes up twice, then back down to my home directory. If a path starts with ~/ then it refers to your home directory. If a path starts with ./ it refers to your current location.
# Home directory
~/
Basic File Operations¶
mkdir creates a new directory with the given name.
# Make directory
mkdir my_first_dir
ls
cp copies a file from one location to another. The example will copy a file containing some text to your home directory.
# Copy: cp source destination
cp /nfs/course/masterdata/tutorials/linux/example_file.txt .
ls
mv moves a file from on location to another. The second mv command example, because the destination is not a directory, actually renames the file. Thus you can move and rename a file with the same command.
# Move or rename: mv source destination
mv ./example_file.txt ./my_first_dir/
ls
ls ./my_first_dir/
mv ./my_first_dir/example_file.txt ./my_first_dir/example_file_renamed.txt
ls ./my_first_dir/
rm removes a file, so use it with care.
# Remove: rm path
rm ./my_first_dir/example_file_renamed.txt
ls ./my_first_dir/
Working with Files¶
In Unix systems there are only really two types of files: text or binary. The file name ending (.txt or .jpg) doesn’t really matter like it does in Windows or MacOS, however it is used to indicate the file type by convention. Some file types you will encounter include:
.txt - A generic text file
.csv - A ‘comma separated values’ file, which is usually a table of data with each line a row and each column separated by a comma
.tsv - A ‘tab separated values’ file, which is the same by separated by tab characters
.fasta or .fa - A fasta formatted sequence file, in which each sequence has a header line starting with ‘>’
.fna - A fasta formatted nucleotide sequence file, usually gene sequences
.faa - A fasta formatted protein sequence file
.sh - A ‘shell script’, which contains terminal commands to run sequentially
.r - An R script, which contains R commands to run
.py - A python script, which contains python commands to run
.gz or .tar.gz - A file that has been compressed using a protocol called ‘gzip’ so that it takes up less space on the disk and transfers over the internet faster
Looking at files¶
cat is a simple command that displays the entire contents of a file directly on the terminal. For large files this can be disastrous, so remember that you can cancel commands in progress with Ctrl+C or cmd+C.
# ConCATenate
cat /nfs/course/masterdata/tutorials/linux/example.fasta
head displays only the first 10 lines of a file directly on the terminal. If you look at the available options for the command, -n x outputs the first x lines instead, and using a negative number outputs the lines except for the last x.
# Show file head
head /nfs/course/masterdata/tutorials/linux/example.fasta
tail displays only the last 10 lines of a file directly on the terminal. It has similar options to head; -n x outputs the last x lines, and using an explicitly positive number +x outputs the lines except for the first x.
# Show file tail
tail /nfs/course/masterdata/tutorials/linux/example.fasta
less is the most versatile and useful way to look at a file in the command line. Instead of showing you the contents of a file directly on the terminal, it ‘opens’ the file to browse. You can use the arrow keys, page up, page down, home, end and the spacebar to navigate the file. Pressing q will quit. A number of useful options exist for the command, such as showing line numbers or displaying without line wrapping. It also has a search feature that we will cover later.
# Browse file
less /nfs/course/masterdata/tutorials/linux/example.fasta
Resources¶
Mastering the terminal is an incredible useful skill for most bioinformatic workflows. We show you only the minimum number commands that are needed for this tutorial. There’re great tutorials if you would like to continue working with the terminal.
Please use the unix cheat sheet as a reference for Linux commands.